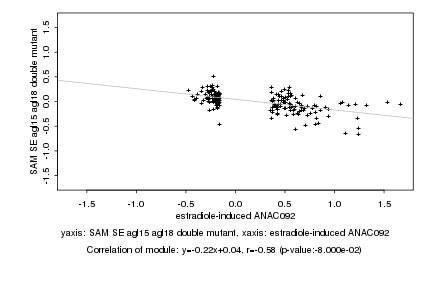
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
estradiole-induced ANAC092 |
Log2 signal ratio
SAM SE agl15 agl18 double mutant |
| 1 |
257944_at |
AT3G21850
|
ASK9, SKP1-like 9, SK9, SKP1-like 9 |
1.662 |
-0.043 |
| 2 |
257943_at |
AT3G21840
|
ASK7, SKP1-like 7, SK7, SKP1-like 7 |
1.531 |
-0.002 |
| 3 |
257942_at |
AT3G21830
|
ASK8, SKP1-like 8, SK8, SKP1-like 8 |
1.316 |
-0.061 |
| 4 |
262454_at |
AT1G11190
|
BFN1, bifunctional nuclease i, ENDO1, ENDONUCLEASE 1 |
1.238 |
-0.662 |
| 5 |
255345_at |
AT4G04460
|
[Saposin-like aspartyl protease family protein] |
1.234 |
-0.532 |
| 6 |
254629_at |
AT4G18425
|
unknown |
1.223 |
-0.330 |
| 7 |
261243_at |
AT1G20180
|
unknown |
1.204 |
-0.055 |
| 8 |
256789_at |
AT3G13672
|
SINA2 |
1.134 |
-0.073 |
| 9 |
263282_at |
AT2G14095
|
unknown |
1.104 |
-0.635 |
| 10 |
252004_at |
AT3G52780
|
ATPAP20, PAP20 |
1.078 |
-0.005 |
| 11 |
262640_at |
AT1G62760
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
1.057 |
-0.028 |
| 12 |
263689_at |
AT1G26820
|
RNS3, ribonuclease 3 |
0.935 |
-0.296 |
| 13 |
267343_at |
AT2G44260
|
unknown |
0.930 |
-0.158 |
| 14 |
253289_at |
AT4G34320
|
unknown |
0.908 |
-0.102 |
| 15 |
262356_at |
AT1G73000
|
PYL3, PYR1-like 3, RCAR13, regulatory components of ABA receptor 13 |
0.855 |
-0.177 |
| 16 |
249454_at |
AT5G39520
|
unknown |
0.854 |
0.121 |
| 17 |
245697_at |
AT5G04200
|
AtMC9, metacaspase 9, AtMCP2f, metacaspase 2f, MC9, metacaspase 9, MCP2f, metacaspase 2f |
0.836 |
-0.428 |
| 18 |
259328_at |
AT3G16440
|
ATMLP-300B, myrosinase-binding protein-like protein-300B, MEE36, maternal effect embryo arrest 36, MLP-300B, myrosinase-binding protein-like protein-300B |
0.813 |
-0.332 |
| 19 |
247965_at |
AT5G56540
|
AGP14, arabinogalactan protein 14, ATAGP14 |
0.813 |
-0.090 |
| 20 |
245982_at |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
0.808 |
-0.215 |
| 21 |
248545_at |
AT5G50260
|
CEP1, cysteine endopeptidase 1 |
0.806 |
-0.457 |
| 22 |
257945_at |
AT3G21860
|
ASK10, SKP1-like 10, SK10, SKP1-like 10 |
0.797 |
-0.066 |
| 23 |
251293_at |
AT3G61930
|
unknown |
0.774 |
-0.133 |
| 24 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
0.749 |
-0.241 |
| 25 |
266021_at |
AT2G05910
|
unknown |
0.725 |
-0.281 |
| 26 |
245705_at |
AT5G04390
|
[C2H2-type zinc finger family protein] |
0.720 |
-0.086 |
| 27 |
266849_at |
AT2G25940
|
ALPHA-VPE, alpha-vacuolar processing enzyme, ALPHAVPE |
0.702 |
-0.466 |
| 28 |
252320_at |
AT3G48580
|
XTH11, xyloglucan endotransglucosylase/hydrolase 11 |
0.696 |
-0.123 |
| 29 |
254823_at |
AT4G12580
|
unknown |
0.679 |
-0.166 |
| 30 |
247657_at |
AT5G59845
|
[Gibberellin-regulated family protein] |
0.674 |
0.127 |
| 31 |
264505_at |
AT1G09380
|
UMAMIT25, Usually multiple acids move in and out Transporters 25 |
0.666 |
-0.111 |
| 32 |
248168_at |
AT5G54570
|
BGLU41, beta glucosidase 41 |
0.659 |
-0.072 |
| 33 |
261385_at |
AT1G05450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.657 |
-0.174 |
| 34 |
254597_at |
AT4G18980
|
AtS40-3, AtS40-3 |
0.646 |
-0.022 |
| 35 |
250535_at |
AT5G08480
|
[VQ motif-containing protein] |
0.637 |
-0.188 |
| 36 |
254956_at |
AT4G10850
|
AtSWEET7, SWEET7 |
0.630 |
-0.243 |
| 37 |
267548_at |
AT2G32660
|
AtRLP22, receptor like protein 22, RLP22, receptor like protein 22 |
0.623 |
-0.011 |
| 38 |
260335_at |
AT1G74000
|
SS3, strictosidine synthase 3 |
0.617 |
-0.240 |
| 39 |
247933_at |
AT5G56980
|
unknown |
0.600 |
0.064 |
| 40 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
0.600 |
-0.098 |
| 41 |
259653_at |
AT1G55240
|
unknown |
0.598 |
-0.549 |
| 42 |
266756_at |
AT2G46950
|
CYP709B2, cytochrome P450, family 709, subfamily B, polypeptide 2 |
0.594 |
-0.150 |
| 43 |
258183_at |
AT3G21550
|
AtDMP2, Arabidopsis thaliana DUF679 domain membrane protein 2, DMP2, DUF679 domain membrane protein 2 |
0.580 |
-0.262 |
| 44 |
252606_at |
AT3G45010
|
scpl48, serine carboxypeptidase-like 48 |
0.576 |
-0.181 |
| 45 |
261441_at |
AT1G28470
|
ANAC010, NAC domain containing protein 10, NAC010, NAC domain containing protein 10, SND3, SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN 3 |
0.573 |
-0.095 |
| 46 |
262940_at |
AT1G79520
|
[Cation efflux family protein] |
0.565 |
-0.116 |
| 47 |
250639_at |
AT5G07560
|
ATGRP20, GRP20, glycine-rich protein 20 |
0.564 |
0.135 |
| 48 |
260048_at |
AT1G73750
|
[Uncharacterised conserved protein UCP031088, alpha/beta hydrolase] |
0.555 |
-0.168 |
| 49 |
253582_at |
AT4G30670
|
[Putative membrane lipoprotein] |
0.554 |
0.163 |
| 50 |
260933_at |
AT1G02470
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.547 |
0.107 |
| 51 |
245436_at |
AT4G16620
|
UMAMIT8, Usually multiple acids move in and out Transporters 8 |
0.547 |
-0.059 |
| 52 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.541 |
0.295 |
| 53 |
262244_at |
AT1G48260
|
CIPK17, CBL-interacting protein kinase 17, SnRK3.21, SNF1-RELATED PROTEIN KINASE 3.21 |
0.541 |
-0.038 |
| 54 |
264144_at |
AT1G79320
|
AtMC6, metacaspase 6, AtMCP2c, metacaspase 2c, MC6, metacaspase 6, MCP2c, metacaspase 2c |
0.537 |
-0.116 |
| 55 |
260261_at |
AT1G68450
|
PDE337, PIGMENT DEFECTIVE 337 |
0.534 |
0.225 |
| 56 |
260135_at |
AT1G66400
|
CML23, calmodulin like 23 |
0.530 |
0.163 |
| 57 |
261366_at |
AT1G53100
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.527 |
0.104 |
| 58 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
0.522 |
0.051 |
| 59 |
254648_at |
AT4G18550
|
AtDSEL, Arabidopsis thaliana DAD1-like seeding establishment-related lipase, DSEL, DAD1-like seeding establishment-related lipase |
0.515 |
-0.280 |
| 60 |
267263_at |
AT2G23110
|
[Late embryogenesis abundant protein, group 6] |
0.501 |
-0.011 |
| 61 |
264652_at |
AT1G08920
|
ESL1, ERD (early response to dehydration) six-like 1 |
0.500 |
-0.124 |
| 62 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
0.498 |
0.084 |
| 63 |
266486_at |
AT2G47950
|
unknown |
0.492 |
-0.131 |
| 64 |
264532_at |
AT1G55740
|
AtSIP1, seed imbibition 1, RS1, raffinose synthase 1, SIP1, seed imbibition 1 |
0.490 |
-0.016 |
| 65 |
259661_at |
AT1G55265
|
unknown |
0.488 |
0.252 |
| 66 |
250208_at |
AT5G14000
|
anac084, NAC domain containing protein 84, NAC084, NAC domain containing protein 84 |
0.481 |
0.039 |
| 67 |
246799_at |
AT5G26940
|
DPD1, defective in pollen organelle DNA degradation1 |
0.481 |
0.062 |
| 68 |
260386_at |
AT1G74010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.478 |
-0.015 |
| 69 |
253676_at |
AT4G29570
|
[Cytidine/deoxycytidylate deaminase family protein] |
0.464 |
0.204 |
| 70 |
248959_at |
AT5G45630
|
unknown |
0.460 |
0.103 |
| 71 |
251019_at |
AT5G02420
|
unknown |
0.457 |
0.028 |
| 72 |
251191_at |
AT3G62590
|
[alpha/beta-Hydrolases superfamily protein] |
0.448 |
-0.081 |
| 73 |
256014_at |
AT1G19200
|
unknown |
0.446 |
-0.049 |
| 74 |
245024_at |
ATCG00120
|
ATPA, ATP synthase subunit alpha |
0.444 |
0.104 |
| 75 |
249794_at |
AT5G23530
|
AtCXE18, carboxyesterase 18, CXE18, carboxyesterase 18 |
0.434 |
-0.044 |
| 76 |
267546_at |
AT2G32680
|
AtRLP23, receptor like protein 23, RLP23, receptor like protein 23 |
0.429 |
0.147 |
| 77 |
246395_at |
AT1G58170
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.429 |
0.037 |
| 78 |
254299_at |
AT4G22920
|
ATNYE1, NON-YELLOWING 1, NYE1, NON-YELLOWING 1, SGR, STAY-GREEN |
0.424 |
-0.157 |
| 79 |
255340_at |
AT4G04490
|
CRK36, cysteine-rich RLK (RECEPTOR-like protein kinase) 36 |
0.423 |
-0.228 |
| 80 |
247312_at |
AT5G63970
|
RGLG3, RING DOMAIN LIGASE 3 |
0.418 |
-0.247 |
| 81 |
264562_at |
AT1G55760
|
[BTB/POZ domain-containing protein] |
0.416 |
-0.151 |
| 82 |
265266_at |
AT2G42890
|
AML2, MEI2-like 2, ML2, MEI2-like 2 |
0.415 |
-0.104 |
| 83 |
249527_at |
AT5G38710
|
[Methylenetetrahydrofolate reductase family protein] |
0.409 |
-0.102 |
| 84 |
264261_at |
AT1G09240
|
ATNAS3, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 3, NAS3, nicotianamine synthase 3 |
0.406 |
0.144 |
| 85 |
264590_at |
AT2G17710
|
unknown |
0.403 |
-0.080 |
| 86 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
0.398 |
-0.080 |
| 87 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
0.391 |
-0.041 |
| 88 |
251060_at |
AT5G01820
|
ATCIPK14, ATSR1, serine/threonine protein kinase 1, CIPK14, CBL-INTERACTING PROTEIN KINASE 14, PKS24, SOS2-like protein kinase 24, SnRK3.15, SNF1-RELATED PROTEIN KINASE 3.15, SR1, serine/threonine protein kinase 1 |
0.391 |
-0.083 |
| 89 |
260887_at |
AT1G29160
|
[Dof-type zinc finger DNA-binding family protein] |
0.382 |
0.079 |
| 90 |
246998_at |
AT5G67370
|
CGLD27, CONSERVED IN THE GREEN LINEAGE AND DIATOMS 27 |
0.379 |
0.030 |
| 91 |
250100_at |
AT5G16570
|
GLN1;4, glutamine synthetase 1;4 |
0.374 |
-0.168 |
| 92 |
252097_at |
AT3G51090
|
unknown |
0.370 |
-0.113 |
| 93 |
253963_at |
AT4G26470
|
[Calcium-binding EF-hand family protein] |
0.365 |
-0.212 |
| 94 |
245025_at |
ATCG00130
|
ATPF |
0.364 |
0.012 |
| 95 |
265913_at |
AT2G25625
|
unknown |
0.360 |
0.192 |
| 96 |
249377_at |
AT5G40690
|
unknown |
0.359 |
-0.344 |
| 97 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
0.357 |
0.294 |
| 98 |
259133_at |
AT3G05400
|
[Major facilitator superfamily protein] |
0.357 |
0.030 |
| 99 |
262478_at |
AT1G11170
|
unknown |
0.353 |
-0.178 |
| 100 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
-0.480 |
0.231 |
| 101 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
-0.434 |
0.113 |
| 102 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
-0.418 |
0.052 |
| 103 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
-0.414 |
0.024 |
| 104 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
-0.403 |
0.064 |
| 105 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
-0.385 |
0.143 |
| 106 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
-0.349 |
0.215 |
| 107 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
-0.348 |
-0.024 |
| 108 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
-0.339 |
0.290 |
| 109 |
259017_at |
AT3G07310
|
unknown |
-0.328 |
0.022 |
| 110 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
-0.307 |
0.145 |
| 111 |
249384_at |
AT5G39890
|
unknown |
-0.300 |
0.067 |
| 112 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
-0.292 |
0.306 |
| 113 |
250152_at |
AT5G15120
|
unknown |
-0.291 |
0.095 |
| 114 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
-0.286 |
0.241 |
| 115 |
263061_at |
AT2G18190
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.283 |
0.078 |
| 116 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
-0.278 |
0.202 |
| 117 |
264246_at |
AT1G60140
|
ATTPS10, trehalose phosphate synthase, TPS10, trehalose phosphate synthase, TPS10, TREHALOSE PHOSPHATE SYNTHASE 10 |
-0.277 |
0.010 |
| 118 |
265428_at |
AT2G20720
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.272 |
0.045 |
| 119 |
245925_at |
AT5G28770
|
AtbZIP63, Arabidopsis thaliana basic leucine zipper 63, BZO2H3 |
-0.267 |
-0.007 |
| 120 |
262505_at |
AT1G21680
|
[DPP6 N-terminal domain-like protein] |
-0.265 |
0.187 |
| 121 |
257153_at |
AT3G27220
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.265 |
0.065 |
| 122 |
249742_at |
AT5G24490
|
[30S ribosomal protein, putative] |
-0.262 |
-0.170 |
| 123 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
-0.262 |
0.118 |
| 124 |
262072_at |
AT1G59590
|
ZCF37 |
-0.259 |
0.305 |
| 125 |
249188_at |
AT5G42830
|
[HXXXD-type acyl-transferase family protein] |
-0.255 |
0.213 |
| 126 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
-0.252 |
0.210 |
| 127 |
254059_at |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
-0.251 |
0.215 |
| 128 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
-0.250 |
0.153 |
| 129 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
-0.247 |
0.238 |
| 130 |
254289_at |
AT4G22980
|
unknown |
-0.240 |
0.074 |
| 131 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
-0.239 |
0.340 |
| 132 |
266291_at |
AT2G29320
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.234 |
0.120 |
| 133 |
254424_at |
AT4G21510
|
AtFBS2, FBS2, F-box stress induced 2 |
-0.234 |
0.303 |
| 134 |
265499_at |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
-0.232 |
0.009 |
| 135 |
257840_at |
AT3G25250
|
AGC2, AGC2-1, AGC2 kinase 1, AtOXI1, OXI1, oxidative signal-inducible1 |
-0.229 |
0.247 |
| 136 |
247922_at |
AT5G57500
|
[Galactosyltransferase family protein] |
-0.228 |
-0.000 |
| 137 |
257621_at |
AT3G20410
|
CPK9, calmodulin-domain protein kinase 9 |
-0.228 |
-0.147 |
| 138 |
261545_at |
AT1G63530
|
unknown |
-0.223 |
0.516 |
| 139 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
-0.220 |
0.218 |
| 140 |
256262_at |
AT3G12150
|
unknown |
-0.219 |
0.209 |
| 141 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
-0.217 |
0.053 |
| 142 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-0.215 |
0.147 |
| 143 |
249847_at |
AT5G23210
|
SCPL34, serine carboxypeptidase-like 34 |
-0.210 |
-0.004 |
| 144 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
-0.210 |
0.035 |
| 145 |
247374_at |
AT5G63190
|
[MA3 domain-containing protein] |
-0.210 |
0.028 |
| 146 |
262124_at |
AT1G59660
|
[Nucleoporin autopeptidase] |
-0.209 |
0.151 |
| 147 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
-0.208 |
0.144 |
| 148 |
245244_at |
AT1G44350
|
ILL6, IAA-leucine resistant (ILR)-like gene 6 |
-0.208 |
0.077 |
| 149 |
257922_at |
AT3G23150
|
ETR2, ethylene response 2 |
-0.206 |
0.120 |
| 150 |
258385_at |
AT3G15510
|
ANAC056, Arabidopsis NAC domain containing protein 56, ATNAC2, NAC domain containing protein 2, NAC2, NAC domain containing protein 2, NARS1, NAC-REGULATED SEED MORPHOLOGY 1 |
-0.204 |
0.142 |
| 151 |
254318_at |
AT4G22530
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.204 |
0.050 |
| 152 |
264836_at |
AT1G03610
|
unknown |
-0.202 |
0.175 |
| 153 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
-0.202 |
0.015 |
| 154 |
257625_at |
AT3G26230
|
CYP71B24, cytochrome P450, family 71, subfamily B, polypeptide 24 |
-0.201 |
0.129 |
| 155 |
260126_at |
AT1G36370
|
SHM7, serine hydroxymethyltransferase 7 |
-0.200 |
-0.014 |
| 156 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
-0.200 |
0.099 |
| 157 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
-0.200 |
0.002 |
| 158 |
247243_at |
AT5G64700
|
UMAMIT21, Usually multiple acids move in and out Transporters 21 |
-0.199 |
0.107 |
| 159 |
255365_at |
AT4G04040
|
MEE51, maternal effect embryo arrest 51 |
-0.196 |
-0.064 |
| 160 |
249112_at |
AT5G43780
|
APS4 |
-0.194 |
0.087 |
| 161 |
252040_at |
AT3G52060
|
AtGnTL, GnTL, beta-1,6-N-acetylglucosaminyl transferase-like |
-0.192 |
0.160 |
| 162 |
253614_at |
AT4G30350
|
SMXL2, SMAX1-like 2 |
-0.192 |
0.022 |
| 163 |
262026_at |
AT1G35670
|
ATCDPK2, calcium-dependent protein kinase 2, ATCPK11, CDPK2, calcium-dependent protein kinase 2, CPK11 |
-0.189 |
-0.042 |
| 164 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
-0.189 |
0.118 |
| 165 |
265457_at |
AT2G46550
|
unknown |
-0.188 |
-0.109 |
| 166 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.188 |
-0.006 |
| 167 |
252740_at |
AT3G43270
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.188 |
0.086 |
| 168 |
258507_at |
AT3G06500
|
A/N-InvC, alkaline/neutral invertase C |
-0.187 |
-0.134 |
| 169 |
267493_at |
AT2G30400
|
ATOFP2, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 2, OFP2, ovate family protein 2 |
-0.187 |
-0.041 |
| 170 |
253404_at |
AT4G32840
|
PFK6, phosphofructokinase 6 |
-0.187 |
0.010 |
| 171 |
246228_at |
AT4G36430
|
[Peroxidase superfamily protein] |
-0.186 |
0.070 |
| 172 |
258189_at |
AT3G17860
|
JAI3, JASMONATE-INSENSITIVE 3, JAZ3, jasmonate-zim-domain protein 3, TIFY6B |
-0.185 |
0.145 |
| 173 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
-0.184 |
0.314 |
| 174 |
266514_at |
AT2G47890
|
[B-box type zinc finger protein with CCT domain] |
-0.180 |
0.102 |
| 175 |
263565_at |
AT2G15390
|
atfut4, FUT4, fucosyltransferase 4 |
-0.180 |
0.174 |
| 176 |
254553_at |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
-0.178 |
-0.040 |
| 177 |
253141_at |
AT4G35440
|
ATCLC-E, CLC-E, chloride channel E, CLCE, CHLORIDE CHANNEL E |
-0.177 |
0.093 |
| 178 |
266983_at |
AT2G39400
|
[alpha/beta-Hydrolases superfamily protein] |
-0.176 |
0.188 |
| 179 |
260014_at |
AT1G68010
|
ATHPR1, HPR, hydroxypyruvate reductase |
-0.176 |
0.050 |
| 180 |
262884_at |
AT1G64720
|
CP5 |
-0.176 |
-0.037 |
| 181 |
248719_at |
AT5G47910
|
ATRBOHD, RBOHD, respiratory burst oxidase homologue D |
-0.176 |
0.039 |
| 182 |
257172_at |
AT3G23700
|
[Nucleic acid-binding proteins superfamily] |
-0.174 |
-0.019 |
| 183 |
261618_at |
AT1G33110
|
[MATE efflux family protein] |
-0.174 |
0.083 |
| 184 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
-0.174 |
0.010 |
| 185 |
247594_at |
AT5G60800
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.174 |
0.043 |
| 186 |
265999_at |
AT2G24100
|
ASG1, ALTERED SEED GERMINATION 1 |
-0.172 |
0.080 |
| 187 |
250248_at |
AT5G13740
|
ZIF1, zinc induced facilitator 1 |
-0.172 |
0.126 |
| 188 |
258114_at |
AT3G14660
|
CYP72A13, cytochrome P450, family 72, subfamily A, polypeptide 13 |
-0.171 |
-0.011 |
| 189 |
253281_at |
AT4G34138
|
UGT73B1, UDP-glucosyl transferase 73B1 |
-0.171 |
0.128 |
| 190 |
256456_at |
AT1G75180
|
[Erythronate-4-phosphate dehydrogenase family protein] |
-0.169 |
0.170 |
| 191 |
248199_at |
AT5G54170
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.167 |
0.030 |
| 192 |
258063_at |
AT3G14620
|
CYP72A8, cytochrome P450, family 72, subfamily A, polypeptide 8 |
-0.167 |
0.000 |
| 193 |
253722_at |
AT4G29190
|
AtC3H49, AtOZF2, Arabidopsis thaliana oxidation-related zinc finger 2, AtTZF3, OZF2, oxidation-related zinc finger 2, TZF3, tandem zinc finger 3 |
-0.165 |
0.178 |
| 194 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
-0.165 |
-0.446 |
| 195 |
264929_at |
AT1G60730
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.165 |
-0.064 |
| 196 |
252199_at |
AT3G50270
|
[HXXXD-type acyl-transferase family protein] |
-0.165 |
0.148 |
| 197 |
252896_at |
AT4G39480
|
CYP96A9, cytochrome P450, family 96, subfamily A, polypeptide 9 |
-0.163 |
0.078 |