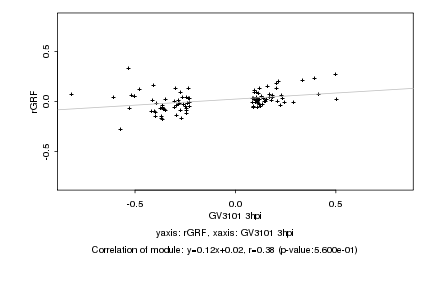
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
GV3101 3hpi |
Log2 signal ratio
rGRF |
| 1 |
267262_at |
AT2G22990
|
SCPL8, SERINE CARBOXYPEPTIDASE-LIKE 8, SNG1, sinapoylglucose 1 |
0.501 |
0.021 |
| 2 |
260783_at |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
0.495 |
0.272 |
| 3 |
251246_at |
AT3G62100
|
IAA30, indole-3-acetic acid inducible 30 |
0.409 |
0.071 |
| 4 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.392 |
0.235 |
| 5 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.329 |
0.220 |
| 6 |
249928_at |
AT5G22250
|
AtCAF1b, CCR4- associated factor 1b, CAF1b, CCR4- associated factor 1b |
0.287 |
-0.002 |
| 7 |
246596_at |
AT5G14740
|
BETA CA2, BETA CARBONIC ANHYDRASE 2, CA18, CARBONIC ANHYDRASE 18, CA2, carbonic anhydrase 2 |
0.241 |
-0.006 |
| 8 |
254093_at |
AT4G25110
|
AtMC2, metacaspase 2, AtMCP1c, metacaspase 1c, MC2, metacaspase 2, MCP1c, metacaspase 1c |
0.234 |
0.031 |
| 9 |
262870_at |
AT1G64710
|
[GroES-like zinc-binding dehydrogenase family protein] |
0.227 |
0.066 |
| 10 |
252092_at |
AT3G51420
|
ATSSL4, STRICTOSIDINE SYNTHASE-LIKE 4, SSL4, strictosidine synthase-like 4 |
0.220 |
-0.031 |
| 11 |
260754_at |
AT1G49000
|
unknown |
0.213 |
0.203 |
| 12 |
247205_at |
AT5G64890
|
PROPEP2, elicitor peptide 2 precursor |
0.208 |
0.002 |
| 13 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
0.201 |
0.134 |
| 14 |
262793_at |
AT1G13110
|
CYP71B7, cytochrome P450, family 71 subfamily B, polypeptide 7 |
0.200 |
0.189 |
| 15 |
262315_at |
AT1G70990
|
[proline-rich family protein] |
0.182 |
0.044 |
| 16 |
254015_at |
AT4G26140
|
BGAL12, beta-galactosidase 12 |
0.180 |
0.069 |
| 17 |
261292_at |
AT1G36940
|
unknown |
0.178 |
0.018 |
| 18 |
257153_at |
AT3G27220
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.169 |
0.047 |
| 19 |
263652_at |
AT1G04330
|
unknown |
0.168 |
0.071 |
| 20 |
251919_at |
AT3G53800
|
Fes1B, Fes1B |
0.156 |
0.156 |
| 21 |
259211_at |
AT3G09020
|
[alpha 1,4-glycosyltransferase family protein] |
0.151 |
0.021 |
| 22 |
251320_at |
AT3G61530
|
PANB2 |
0.149 |
0.006 |
| 23 |
247278_at |
AT5G64380
|
[Inositol monophosphatase family protein] |
0.140 |
0.008 |
| 24 |
249327_at |
AT5G40890
|
ATCLC-A, chloride channel A, ATCLCA, CLC-A, chloride channel A, CLC-A, CHLORIDE CHANNEL-A, CLCA, CHLORIDE CHANNEL A |
0.138 |
0.032 |
| 25 |
262288_at |
AT1G70760
|
CRR23, CHLORORESPIRATORY REDUCTION 23, NdhL, NADH dehydrogenase-like complex L |
0.132 |
-0.024 |
| 26 |
262552_at |
AT1G31350
|
KUF1, KAR-UP F-box 1 |
0.129 |
0.055 |
| 27 |
252441_at |
AT3G46780
|
PTAC16, plastid transcriptionally active 16 |
0.121 |
-0.040 |
| 28 |
246019_at |
AT5G10690
|
[pentatricopeptide (PPR) repeat-containing protein / CBS domain-containing protein] |
0.119 |
-0.020 |
| 29 |
263565_at |
AT2G15390
|
atfut4, FUT4, fucosyltransferase 4 |
0.118 |
0.027 |
| 30 |
254256_at |
AT4G23180
|
CRK10, cysteine-rich RLK (RECEPTOR-like protein kinase) 10, RLK4 |
0.116 |
0.135 |
| 31 |
250498_at |
AT5G09660
|
PMDH2, peroxisomal NAD-malate dehydrogenase 2 |
0.115 |
-0.025 |
| 32 |
250948_at |
AT5G03490
|
[UDP-Glycosyltransferase superfamily protein] |
0.114 |
-0.011 |
| 33 |
252387_at |
AT3G47800
|
[Galactose mutarotase-like superfamily protein] |
0.113 |
0.090 |
| 34 |
266860_at |
AT2G26870
|
NPC2, non-specific phospholipase C2 |
0.112 |
0.027 |
| 35 |
256262_at |
AT3G12150
|
unknown |
0.108 |
0.028 |
| 36 |
253065_at |
AT4G37740
|
AtGRF2, growth-regulating factor 2, GRF2, growth-regulating factor 2 |
0.107 |
0.010 |
| 37 |
260206_at |
AT1G70740
|
[Protein kinase superfamily protein] |
0.107 |
0.013 |
| 38 |
259499_at |
AT1G15730
|
[Cobalamin biosynthesis CobW-like protein] |
0.107 |
-0.050 |
| 39 |
246028_at |
AT5G21170
|
5'-AMP-activated protein kinase beta-2 subunit protein |
0.103 |
0.100 |
| 40 |
261084_at |
AT1G07440
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.102 |
0.047 |
| 41 |
259484_at |
AT1G12580
|
PEPKR1, phosphoenolpyruvate carboxylase-related kinase 1 |
0.099 |
-0.008 |
| 42 |
253750_at |
AT4G29000
|
[Tesmin/TSO1-like CXC domain-containing protein] |
0.098 |
0.012 |
| 43 |
267490_at |
AT2G19130
|
[S-locus lectin protein kinase family protein] |
0.093 |
0.091 |
| 44 |
256975_at |
AT3G21000
|
[Gag-Pol-related retrotransposon family protein] |
0.093 |
0.027 |
| 45 |
255617_at |
AT4G01330
|
[Protein kinase superfamily protein] |
0.091 |
0.111 |
| 46 |
246906_at |
AT5G25475
|
[AP2/B3-like transcriptional factor family protein] |
0.091 |
0.021 |
| 47 |
251914_at |
AT3G53930
|
[Protein kinase superfamily protein] |
0.089 |
-0.059 |
| 48 |
260733_at |
AT1G17640
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.089 |
-0.058 |
| 49 |
259694_at |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
0.088 |
0.032 |
| 50 |
248788_at |
AT5G47430
|
[DWNN domain, a CCHC-type zinc finger] |
0.087 |
-0.040 |
| 51 |
248302_at |
AT5G53160
|
PYL8, PYR1-like 8, RCAR3, regulatory components of ABA receptor 3 |
0.080 |
0.033 |
| 52 |
267562_at |
AT2G39670
|
[Radical SAM superfamily protein] |
0.080 |
-0.004 |
| 53 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
-0.820 |
0.077 |
| 54 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
-0.612 |
0.045 |
| 55 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
-0.576 |
-0.277 |
| 56 |
259802_at |
AT1G72260
|
THI2.1, thionin 2.1, THI2.1.1 |
-0.534 |
0.335 |
| 57 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
-0.530 |
-0.064 |
| 58 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
-0.522 |
0.064 |
| 59 |
266883_at |
AT2G44810
|
DAD1, DEFECTIVE ANTHER DEHISCENCE 1 |
-0.506 |
0.052 |
| 60 |
251513_at |
AT3G59220
|
ATPIRIN1, PRN, pirin, PRN1 |
-0.505 |
0.051 |
| 61 |
256933_at |
AT3G22600
|
LTPG5, glycosylphosphatidylinositol-anchored lipid protein transfer 5 |
-0.479 |
0.130 |
| 62 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
-0.419 |
-0.096 |
| 63 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
-0.415 |
0.019 |
| 64 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
-0.413 |
0.164 |
| 65 |
250558_at |
AT5G07990
|
CYP75B1, CYTOCHROME P450 75B1, D501, TT7, TRANSPARENT TESTA 7 |
-0.408 |
-0.092 |
| 66 |
255056_at |
AT4G09820
|
AtTT8, BHLH42, TT8, TRANSPARENT TESTA 8 |
-0.403 |
-0.143 |
| 67 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
-0.402 |
-0.105 |
| 68 |
244970_at |
ATCG00660
|
RPL20, ribosomal protein L20 |
-0.397 |
-0.017 |
| 69 |
264271_at |
AT1G60270
|
BGLU6, beta glucosidase 6 |
-0.375 |
-0.060 |
| 70 |
250420_at |
AT5G11260
|
HY5, ELONGATED HYPOCOTYL 5, TED 5, REVERSAL OF THE DET PHENOTYPE 5 |
-0.372 |
-0.160 |
| 71 |
246966_at |
AT5G24850
|
CRY3, cryptochrome 3 |
-0.369 |
-0.143 |
| 72 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
-0.368 |
-0.053 |
| 73 |
261907_at |
AT1G65060
|
4CL3, 4-coumarate:CoA ligase 3 |
-0.368 |
-0.171 |
| 74 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.364 |
-0.030 |
| 75 |
256924_at |
AT3G29590
|
AT5MAT |
-0.363 |
-0.071 |
| 76 |
252073_at |
AT3G51750
|
unknown |
-0.358 |
-0.072 |
| 77 |
249063_at |
AT5G44110
|
ABCI21, ATP-binding cassette A21, ATNAP2, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN, ATPOP1, POP1 |
-0.352 |
-0.086 |
| 78 |
259793_at |
AT1G64380
|
[Integrase-type DNA-binding superfamily protein] |
-0.352 |
0.027 |
| 79 |
253293_at |
AT4G33905
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
-0.307 |
-0.057 |
| 80 |
251858_at |
AT3G54820
|
PIP2;5, plasma membrane intrinsic protein 2;5, PIP2D, PLASMA MEMBRANE INTRINSIC PROTEIN 2D |
-0.306 |
0.010 |
| 81 |
249971_at |
AT5G19110
|
[Eukaryotic aspartyl protease family protein] |
-0.303 |
0.134 |
| 82 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-0.298 |
-0.135 |
| 83 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.297 |
-0.039 |
| 84 |
261785_at |
AT1G08230
|
[Transmembrane amino acid transporter family protein] |
-0.286 |
0.015 |
| 85 |
265544_at |
AT2G28260
|
ATCNGC15, cyclic nucleotide-gated channel 15, CNGC15, cyclic nucleotide-gated channel 15 |
-0.284 |
-0.021 |
| 86 |
248855_at |
AT5G46590
|
anac096, NAC domain containing protein 96, NAC096, NAC domain containing protein 96 |
-0.283 |
-0.014 |
| 87 |
259383_at |
AT3G16470
|
JAL35, jacalin-related lectin 35, JR1, JASMONATE RESPONSIVE 1 |
-0.278 |
0.097 |
| 88 |
261016_at |
AT1G26560
|
BGLU40, beta glucosidase 40 |
-0.278 |
-0.085 |
| 89 |
263796_at |
AT2G24540
|
AFR, ATTENUATED FAR-RED RESPONSE |
-0.271 |
-0.164 |
| 90 |
246235_at |
AT4G36830
|
HOS3-1 |
-0.266 |
0.047 |
| 91 |
263789_at |
AT2G24560
|
[GDSL-like Lipase/Acylhydrolase family protein] |
-0.262 |
-0.028 |
| 92 |
249118_at |
AT5G43870
|
[FUNCTIONS IN: phosphoinositide binding] |
-0.251 |
-0.048 |
| 93 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
-0.247 |
-0.086 |
| 94 |
253880_at |
AT4G27590
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.247 |
-0.115 |
| 95 |
247827_at |
AT5G58500
|
LSH5, LIGHT SENSITIVE HYPOCOTYLS 5 |
-0.244 |
0.042 |
| 96 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
-0.244 |
-0.060 |
| 97 |
250690_at |
AT5G06530
|
ABCG22, ATP-binding cassette G22, AtABCG22, Arabidopsis thaliana ATP-binding cassette G22 |
-0.243 |
-0.012 |
| 98 |
258356_at |
AT3G14340
|
unknown |
-0.240 |
-0.016 |
| 99 |
250826_at |
AT5G05220
|
unknown |
-0.238 |
0.140 |
| 100 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
-0.237 |
0.033 |
| 101 |
247904_at |
AT5G57390
|
AIL5, AINTEGUMENTA-like 5, CHO1, CHOTTO 1, EMK, EMBRYOMAKER, PLT5, PLETHORA 5 |
-0.230 |
0.033 |
| 102 |
250413_at |
AT5G11160
|
APT5, adenine phosphoribosyltransferase 5 |
-0.230 |
-0.046 |
| 103 |
247776_at |
AT5G58700
|
ATPLC4, PLC4, phosphatidylinositol-speciwc phospholipase C4 |
-0.229 |
-0.000 |