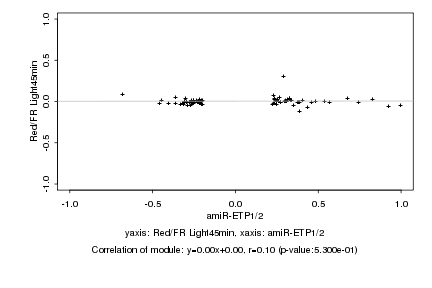
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
amiR-ETP1/2 |
Log2 signal ratio
Red/FR Light45min |
| 1 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.994 |
-0.040 |
| 2 |
253707_at |
AT4G29200
|
[Beta-galactosidase related protein] |
0.921 |
-0.056 |
| 3 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.826 |
0.028 |
| 4 |
245766_at |
AT1G33580
|
[CACTA-like transposase family (Tnp2/En/Spm), has a 5.5e-96 P-value blast match to gb|AAG52024.1|AC022456_5 Tam1-homologous transposon protein TNP2, putative] |
0.741 |
-0.007 |
| 5 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.673 |
0.044 |
| 6 |
254110_at |
AT4G25260
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.562 |
-0.008 |
| 7 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.533 |
0.007 |
| 8 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.483 |
0.004 |
| 9 |
252474_at |
AT3G46620
|
unknown |
0.454 |
-0.004 |
| 10 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.432 |
-0.065 |
| 11 |
250891_at |
AT5G04530
|
KCS19, 3-ketoacyl-CoA synthase 19 |
0.401 |
0.024 |
| 12 |
262641_at |
AT1G62730
|
[Terpenoid synthases superfamily protein] |
0.386 |
-0.120 |
| 13 |
253836_at |
AT4G27840
|
[SNARE-like superfamily protein] |
0.385 |
-0.003 |
| 14 |
265063_at |
AT1G61500
|
[S-locus lectin protein kinase family protein] |
0.375 |
-0.007 |
| 15 |
259850_at |
AT1G72240
|
unknown |
0.370 |
-0.011 |
| 16 |
259535_at |
AT1G12280
|
SUMM2, suppressor of mkk1 mkk2 2 |
0.350 |
-0.037 |
| 17 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.337 |
0.015 |
| 18 |
267080_at |
AT2G41190
|
[Transmembrane amino acid transporter family protein] |
0.330 |
0.024 |
| 19 |
253122_at |
AT4G35987
|
CaM KMT, calmodulin N-methyltransferase |
0.325 |
0.041 |
| 20 |
263043_at |
AT1G23350
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.321 |
0.045 |
| 21 |
256596_at |
AT3G28540
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.309 |
0.026 |
| 22 |
263617_at |
AT2G04670
|
[gypsy-like retrotransposon family, has a 1.2e-313 P-value blast match to GB:AAD27547 polyprotein (Gypsy_Ty3-element) (Oryza sativa subsp. indica)] |
0.307 |
0.008 |
| 23 |
263772_at |
AT2G06255
|
ELF4-L3, ELF4-like 3 |
0.304 |
0.005 |
| 24 |
256021_at |
AT1G58270
|
ZW9 |
0.302 |
0.010 |
| 25 |
257396_at |
AT2G20875
|
EPF1, EPIDERMAL PATTERNING FACTOR 1 |
0.297 |
0.018 |
| 26 |
254608_at |
AT4G18910
|
ATNLM2, NOD26-LIKE INTRINSIC PROTEIN 2, NIP1;2, NOD26-like intrinsic protein 1;2, NLM2, NOD26-LIKE INTRINSIC PROTEIN 2 |
0.292 |
0.010 |
| 27 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
0.288 |
0.310 |
| 28 |
249682_at |
AT5G36080
|
unknown |
0.271 |
-0.008 |
| 29 |
260903_at |
AT1G02460
|
[Pectin lyase-like superfamily protein] |
0.267 |
-0.007 |
| 30 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
0.262 |
0.053 |
| 31 |
256599_at |
AT3G14760
|
unknown |
0.259 |
0.002 |
| 32 |
266415_at |
AT2G38530
|
cdf3, cell growth defect factor-3, LP2, LTP2, lipid transfer protein 2 |
0.258 |
0.003 |
| 33 |
260345_at |
AT1G69270
|
AtRPK1, RPK1, receptor-like protein kinase 1 |
0.252 |
0.026 |
| 34 |
266731_at |
AT2G03260
|
[EXS (ERD1/XPR1/SYG1) family protein] |
0.247 |
0.008 |
| 35 |
250176_at |
AT5G14400
|
CYP724A1, cytochrome P450, family 724, subfamily A, polypeptide 1 |
0.244 |
-0.024 |
| 36 |
248321_at |
AT5G52740
|
[Copper transport protein family] |
0.238 |
0.014 |
| 37 |
252649_at |
AT3G44680
|
AtHDA9, HDA09, HDA9, histone deacetylase 9 |
0.234 |
0.043 |
| 38 |
259606_at |
AT1G27920
|
MAP65-8, microtubule-associated protein 65-8 |
0.234 |
0.025 |
| 39 |
257334_at |
ATMG01370
|
ORF111D |
0.230 |
-0.022 |
| 40 |
248341_at |
AT5G52220
|
unknown |
0.225 |
0.082 |
| 41 |
265267_at |
AT2G42920
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.225 |
-0.012 |
| 42 |
265140_at |
AT1G51320
|
[F-box and associated interaction domains-containing protein] |
0.222 |
-0.028 |
| 43 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.684 |
0.086 |
| 44 |
245381_at |
AT4G17785
|
MYB39, myb domain protein 39 |
-0.462 |
-0.018 |
| 45 |
260417_at |
AT1G69770
|
CMT3, chromomethylase 3 |
-0.450 |
0.015 |
| 46 |
265588_at |
AT2G19970
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.407 |
-0.016 |
| 47 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
-0.367 |
-0.020 |
| 48 |
247800_at |
AT5G58570
|
unknown |
-0.363 |
0.053 |
| 49 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-0.335 |
-0.028 |
| 50 |
254828_at |
AT4G12550
|
AIR1, Auxin-Induced in Root cultures 1 |
-0.321 |
-0.014 |
| 51 |
260256_at |
AT1G74350
|
[Intron maturase, type II family protein] |
-0.319 |
-0.030 |
| 52 |
265341_at |
AT2G18360
|
[alpha/beta-Hydrolases superfamily protein] |
-0.310 |
-0.008 |
| 53 |
262045_at |
AT1G80240
|
DGR1, DUF642 L-GalL responsive gene 1 |
-0.308 |
0.001 |
| 54 |
247438_at |
AT5G62460
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-0.308 |
0.008 |
| 55 |
250471_at |
AT5G10170
|
ATMIPS3, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 3, MIPS3, myo-inositol-1-phosphate synthase 3 |
-0.307 |
0.041 |
| 56 |
261671_at |
AT1G18340
|
[basal transcription factor complex subunit-related] |
-0.298 |
0.002 |
| 57 |
246450_at |
AT5G16820
|
ATHSF3, ARABIDOPSIS HEAT SHOCK FACTOR 3, ATHSFA1B, ARABIDOPSIS THALIANA CLASS A HEAT SHOCK FACTOR 1B, HSF3, heat shock factor 3, HSFA1B, CLASS A HEAT SHOCK FACTOR 1B |
-0.296 |
-0.011 |
| 58 |
247145_at |
AT5G65600
|
LecRK-IX.2, L-type lectin receptor kinase IX.2 |
-0.291 |
-0.037 |
| 59 |
249378_at |
AT5G40450
|
unknown |
-0.283 |
-0.009 |
| 60 |
265771_at |
AT2G48030
|
[DNAse I-like superfamily protein] |
-0.278 |
-0.005 |
| 61 |
263259_at |
AT1G10560
|
ATPUB18, ARABIDOPSIS THALIANA PLANT U-BOX 18, PUB18, plant U-box 18 |
-0.272 |
-0.040 |
| 62 |
253613_at |
AT4G30320
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.271 |
-0.031 |
| 63 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
-0.267 |
0.002 |
| 64 |
246475_at |
AT5G16720
|
MyoB3, myosin binding protein 3 |
-0.266 |
0.018 |
| 65 |
246440_at |
AT5G17650
|
[glycine/proline-rich protein] |
-0.259 |
-0.018 |
| 66 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
-0.258 |
0.021 |
| 67 |
247867_at |
AT5G57630
|
CIPK21, CBL-interacting protein kinase 21, SnRK3.4, SNF1-RELATED PROTEIN KINASE 3.4 |
-0.256 |
-0.009 |
| 68 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
-0.255 |
0.003 |
| 69 |
258537_at |
AT3G04210
|
[Disease resistance protein (TIR-NBS class)] |
-0.254 |
0.016 |
| 70 |
246653_at |
AT5G35200
|
[ENTH/ANTH/VHS superfamily protein] |
-0.250 |
-0.008 |
| 71 |
247167_at |
AT5G65850
|
[F-box and associated interaction domains-containing protein] |
-0.241 |
0.015 |
| 72 |
264744_at |
AT1G62050
|
[Ankyrin repeat family protein] |
-0.231 |
-0.002 |
| 73 |
247348_at |
AT5G63810
|
AtBGAL10, Arabidopsis thaliana beta-galactosidase 10, BGAL10, beta-galactosidase 10 |
-0.224 |
0.012 |
| 74 |
266929_at |
AT2G45850
|
AHL9, AT-hook motif nuclear localized protein 9 |
-0.218 |
-0.022 |
| 75 |
265785_at |
AT2G07290
|
unknown |
-0.218 |
-0.015 |
| 76 |
257515_at |
AT1G13290
|
DOT5, DEFECTIVELY ORGANIZED TRIBUTARIES 5, WIP6, WIP domain protein 6 |
-0.217 |
0.025 |
| 77 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
-0.215 |
0.009 |
| 78 |
254518_at |
AT4G19910
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.211 |
0.004 |
| 79 |
262731_at |
AT1G16420
|
ATMC8, ARABIDOPSIS THALIANA METACASPASE 8, AtMCP2e, metacaspase 2e, MC8, metacaspase 8, MCP2e, metacaspase 2e |
-0.210 |
0.012 |
| 80 |
250223_at |
AT5G14070
|
ROXY2 |
-0.205 |
0.023 |
| 81 |
254972_at |
AT4G10440
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.205 |
-0.027 |
| 82 |
260887_at |
AT1G29160
|
[Dof-type zinc finger DNA-binding family protein] |
-0.202 |
-0.032 |
| 83 |
263498_at |
AT2G42610
|
LSH10, LIGHT SENSITIVE HYPOCOTYLS 10 |
-0.200 |
0.019 |