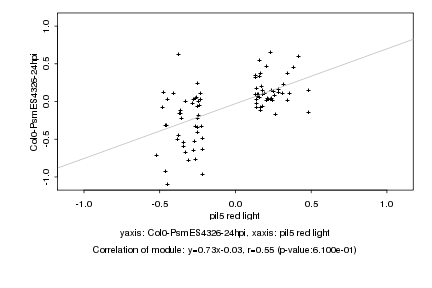
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
pil5 red light |
Log2 signal ratio
Col0-PsmES4326-24hpi |
| 1 |
249214_at |
AT5G42720
|
[Glycosyl hydrolase family 17 protein] |
0.479 |
-0.145 |
| 2 |
261749_at |
AT1G76180
|
ERD14, EARLY RESPONSE TO DEHYDRATION 14 |
0.476 |
0.159 |
| 3 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.416 |
0.610 |
| 4 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
0.379 |
0.464 |
| 5 |
261889_at |
AT1G80810
|
[Tudor/PWWP/MBT superfamily protein] |
0.352 |
0.107 |
| 6 |
264522_at |
AT1G10050
|
[glycosyl hydrolase family 10 protein / carbohydrate-binding domain-containing protein] |
0.343 |
0.373 |
| 7 |
259224_at |
AT3G07800
|
AtTK1a, TK1a, thymidine kinase 1a |
0.341 |
0.021 |
| 8 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
0.317 |
0.230 |
| 9 |
264720_at |
AT1G70080
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.310 |
0.110 |
| 10 |
264617_at |
AT2G17660
|
[RPM1-interacting protein 4 (RIN4) family protein] |
0.282 |
0.163 |
| 11 |
259169_at |
AT3G03520
|
NPC3, non-specific phospholipase C3 |
0.278 |
0.126 |
| 12 |
250372_at |
AT5G11460
|
unknown |
0.261 |
-0.165 |
| 13 |
258879_at |
AT3G03270
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.256 |
0.089 |
| 14 |
245456_at |
AT4G16950
|
RPP5, RECOGNITION OF PERONOSPORA PARASITICA 5 |
0.245 |
0.135 |
| 15 |
257574_at |
AT3G20710
|
[F-box family protein] |
0.242 |
0.021 |
| 16 |
266511_at |
AT2G47680
|
[zinc finger (CCCH type) helicase family protein] |
0.234 |
0.150 |
| 17 |
264604_at |
AT1G04650
|
unknown |
0.233 |
0.044 |
| 18 |
256188_at |
AT1G30160
|
unknown |
0.231 |
0.031 |
| 19 |
257154_at |
AT3G27210
|
unknown |
0.231 |
0.659 |
| 20 |
266008_at |
AT2G37390
|
NAKR2, SODIUM POTASSIUM ROOT DEFECTIVE 2 |
0.215 |
0.028 |
| 21 |
260668_at |
AT1G19530
|
unknown |
0.206 |
0.050 |
| 22 |
245903_at |
AT5G11100
|
ATSYTD, NTMC2T2.2, NTMC2TYPE2.2, SYT4, synaptotagmin 4, SYTD |
0.202 |
0.018 |
| 23 |
259403_at |
AT1G17745
|
PGDH, 3-phosphoglycerate dehydrogenase, PGDH2, phosphoglycerate dehydrogenase 2 |
0.199 |
0.473 |
| 24 |
261643_at |
AT1G27720
|
TAF4, TBP-associated factor 4, TAF4B, TBP-associated factor 4B |
0.191 |
0.114 |
| 25 |
258170_at |
AT3G21600
|
[Senescence/dehydration-associated protein-related] |
0.178 |
0.155 |
| 26 |
249736_at |
AT5G24460
|
unknown |
0.178 |
0.101 |
| 27 |
250186_at |
AT5G14500
|
[aldose 1-epimerase family protein] |
0.176 |
-0.057 |
| 28 |
259186_at |
AT3G01590
|
[Galactose mutarotase-like superfamily protein] |
0.169 |
0.212 |
| 29 |
247974_at |
AT5G56780
|
ATET2, ARABIDOPSIS EFFECTOR OF TRANSCRIPTION2, ET2, effector of transcription2 |
0.165 |
-0.115 |
| 30 |
259852_at |
AT1G72280
|
AERO1, endoplasmic reticulum oxidoreductins 1, ERO1, endoplasmic reticulum oxidoreductins 1 |
0.164 |
0.373 |
| 31 |
264870_at |
AT1G24210
|
[Paired amphipathic helix (PAH2) superfamily protein] |
0.163 |
-0.067 |
| 32 |
249237_at |
AT5G42050
|
[DCD (Development and Cell Death) domain protein] |
0.158 |
0.333 |
| 33 |
258117_at |
AT3G14700
|
[SART-1 family] |
0.156 |
0.550 |
| 34 |
257799_at |
AT3G15890
|
[Protein kinase superfamily protein] |
0.154 |
0.057 |
| 35 |
256138_at |
AT1G48670
|
[auxin-responsive GH3 family protein] |
0.147 |
0.102 |
| 36 |
259606_at |
AT1G27920
|
MAP65-8, microtubule-associated protein 65-8 |
0.143 |
0.079 |
| 37 |
247333_at |
AT5G63600
|
ATFLS5, FLS5, flavonol synthase 5 |
0.143 |
0.106 |
| 38 |
245276_at |
AT4G16780
|
ATHB-2, homeobox protein 2, ATHB2, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2, HAT4, HB-2, homeobox protein 2 |
0.139 |
-0.016 |
| 39 |
253038_at |
AT4G37790
|
HAT22 |
0.139 |
0.185 |
| 40 |
259087_at |
AT3G04980
|
[DNAJ heat shock N-terminal domain-containing protein] |
0.137 |
0.032 |
| 41 |
261629_at |
AT1G49980
|
[DNA/RNA polymerases superfamily protein] |
0.135 |
-0.076 |
| 42 |
264287_at |
AT1G61930
|
unknown |
0.131 |
0.100 |
| 43 |
258436_at |
AT3G16720
|
ATL2, TOXICOS EN LEVADURA 2, TL2, TOXICOS EN LEVADURA 2 |
0.130 |
0.330 |
| 44 |
266566_at |
AT2G24040
|
[Low temperature and salt responsive protein family] |
0.129 |
0.350 |
| 45 |
255302_at |
AT4G04830
|
ATMSRB5, methionine sulfoxide reductase B5, MSRB5, methionine sulfoxide reductase B5 |
-0.522 |
-0.707 |
| 46 |
263207_at |
AT1G10550
|
XET, XYLOGLUCAN:XYLOGLUCOSYL TRANSFERASE 33, XTH33, xyloglucan:xyloglucosyl transferase 33 |
-0.485 |
-0.066 |
| 47 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
-0.480 |
0.125 |
| 48 |
256125_at |
AT1G18250
|
ATLP-1 |
-0.466 |
-0.317 |
| 49 |
266215_at |
AT2G06850
|
EXGT-A1, endoxyloglucan transferase A1, EXT, ENDOXYLOGLUCAN TRANSFERASE, XTH4, xyloglucan endotransglucosylase/hydrolase 4 |
-0.462 |
-0.919 |
| 50 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-0.460 |
-0.308 |
| 51 |
267389_at |
AT2G44460
|
BGLU28, beta glucosidase 28 |
-0.449 |
0.035 |
| 52 |
264371_at |
AT1G12090
|
ELP, extensin-like protein |
-0.449 |
-1.087 |
| 53 |
263829_at |
AT2G40435
|
unknown |
-0.411 |
0.116 |
| 54 |
251899_at |
AT3G54400
|
[Eukaryotic aspartyl protease family protein] |
-0.389 |
-0.491 |
| 55 |
254387_at |
AT4G21850
|
ATMSRB9, methionine sulfoxide reductase B9, MSRB9, methionine sulfoxide reductase B9 |
-0.379 |
0.635 |
| 56 |
252624_at |
AT3G44735
|
ATPSK3, PHYTOSULFOKINE 3 PRECURSOR, PSK1, PSK3, PHYTOSULFOKINE 3 PRECURSOR |
-0.379 |
-0.440 |
| 57 |
261075_at |
AT1G07280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.374 |
-0.147 |
| 58 |
248372_at |
AT5G51850
|
TRM24, TON1 Recruiting Motif 24 |
-0.369 |
-0.152 |
| 59 |
256021_at |
AT1G58270
|
ZW9 |
-0.363 |
-0.107 |
| 60 |
256302_at |
AT1G69526
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.362 |
-0.215 |
| 61 |
261975_at |
AT1G64640
|
AtENODL8, ENODL8, early nodulin-like protein 8 |
-0.349 |
-0.594 |
| 62 |
261949_at |
AT1G64670
|
BDG1, BODYGUARD1, CED1, 9-CIS EPOXYCAROTENOID DIOXYGENASE DEFECTIVE 1 |
-0.347 |
-0.532 |
| 63 |
256275_at |
AT3G12110
|
ACT11, actin-11 |
-0.331 |
-0.675 |
| 64 |
247035_at |
AT5G67110
|
ALC, ALCATRAZ |
-0.330 |
0.013 |
| 65 |
248921_at |
AT5G45950
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.315 |
-0.780 |
| 66 |
258299_at |
AT3G23410
|
ATFAO3, ARABIDOPSIS FATTY ALCOHOL OXIDASE 3, FAO3, fatty alcohol oxidase 3 |
-0.287 |
-0.023 |
| 67 |
260541_at |
AT2G43530
|
[Scorpion toxin-like knottin superfamily protein] |
-0.282 |
0.029 |
| 68 |
262414_at |
AT1G49430
|
LACS2, long-chain acyl-CoA synthetase 2, LRD2, LATERAL ROOT DEVELOPMENT 2 |
-0.278 |
-0.646 |
| 69 |
248246_at |
AT5G53200
|
TRY, TRIPTYCHON |
-0.274 |
-0.526 |
| 70 |
253073_at |
AT4G37410
|
CYP81F4, cytochrome P450, family 81, subfamily F, polypeptide 4 |
-0.269 |
0.052 |
| 71 |
262969_at |
AT1G75710
|
[C2H2-like zinc finger protein] |
-0.267 |
-0.322 |
| 72 |
245196_at |
AT1G67750
|
[Pectate lyase family protein] |
-0.266 |
-0.760 |
| 73 |
252623_at |
AT3G45320
|
unknown |
-0.263 |
0.060 |
| 74 |
254363_at |
AT4G22010
|
sks4, SKU5 similar 4 |
-0.251 |
-0.406 |
| 75 |
250875_at |
AT5G04020
|
[calmodulin binding] |
-0.251 |
0.251 |
| 76 |
256528_at |
AT1G66140
|
ZFP4, zinc finger protein 4 |
-0.251 |
-0.224 |
| 77 |
246002_at |
AT5G20740
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.251 |
-0.341 |
| 78 |
253317_at |
AT4G33960
|
unknown |
-0.251 |
-0.065 |
| 79 |
247786_at |
AT5G58600
|
PMR5, POWDERY MILDEW RESISTANT 5, TBL44, TRICHOME BIREFRINGENCE-LIKE 44 |
-0.249 |
0.003 |
| 80 |
263979_at |
AT2G42840
|
PDF1, protodermal factor 1 |
-0.245 |
-0.173 |
| 81 |
251846_at |
AT3G54560
|
HTA11, histone H2A 11 |
-0.238 |
-0.040 |
| 82 |
267173_at |
AT2G37560
|
ATORC2, ORIGIN RECOGNITION COMPLEX SECOND LARGEST SUBUNIT 2, ORC2, origin recognition complex second largest subunit 2 |
-0.237 |
0.028 |
| 83 |
255751_at |
AT1G31950
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.233 |
0.112 |
| 84 |
256516_at |
AT1G66150
|
TMK1, transmembrane kinase 1 |
-0.227 |
-0.327 |
| 85 |
261016_at |
AT1G26560
|
BGLU40, beta glucosidase 40 |
-0.223 |
-0.488 |
| 86 |
252943_at |
AT4G39330
|
ATCAD9, CAD9, cinnamyl alcohol dehydrogenase 9 |
-0.222 |
-0.956 |
| 87 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
-0.220 |
-0.636 |