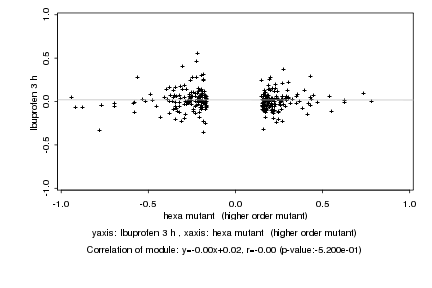
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
hexa mutant (higher order mutant) |
Log2 signal ratio
Ibuprofen 3 h |
| 1 |
250135_at |
AT5G15360
|
unknown |
0.779 |
0.006 |
| 2 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
0.733 |
0.093 |
| 3 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
0.624 |
0.017 |
| 4 |
262421_at |
AT1G50290
|
unknown |
0.621 |
-0.009 |
| 5 |
246552_at |
AT5G15420
|
unknown |
0.551 |
-0.109 |
| 6 |
253999_at |
AT4G26200
|
ACCS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ACS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ATACS7 |
0.535 |
0.058 |
| 7 |
256969_at |
AT3G21080
|
[ABC transporter-related] |
0.468 |
-0.008 |
| 8 |
249955_at |
AT5G18840
|
[Major facilitator superfamily protein] |
0.443 |
0.078 |
| 9 |
253532_at |
AT4G31570
|
unknown |
0.433 |
0.028 |
| 10 |
254314_at |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
0.430 |
0.299 |
| 11 |
249843_at |
AT5G23570
|
ATSGS3, SUPPRESSOR OF GENE SILENCING 3, SGS3, SUPPRESSOR OF GENE SILENCING 3 |
0.430 |
-0.041 |
| 12 |
246168_at |
AT5G32460
|
[Transcriptional factor B3 family protein] |
0.426 |
0.048 |
| 13 |
264872_at |
AT1G24260
|
AGL9, AGAMOUS-like 9, SEP3, SEPALLATA3 |
0.416 |
-0.018 |
| 14 |
248895_at |
AT5G46330
|
FLS2, FLAGELLIN-SENSITIVE 2 |
0.412 |
-0.139 |
| 15 |
249970_at |
AT5G19100
|
[Eukaryotic aspartyl protease family protein] |
0.396 |
0.130 |
| 16 |
248197_at |
AT5G54190
|
PORA, protochlorophyllide oxidoreductase A |
0.380 |
-0.078 |
| 17 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
0.367 |
0.003 |
| 18 |
246814_at |
AT5G27200
|
ACP5, acyl carrier protein 5 |
0.353 |
0.092 |
| 19 |
254350_at |
AT4G22280
|
[F-box/RNI-like superfamily protein] |
0.350 |
-0.017 |
| 20 |
257636_at |
AT3G26200
|
CYP71B22, cytochrome P450, family 71, subfamily B, polypeptide 22 |
0.348 |
0.058 |
| 21 |
256976_at |
AT3G21020
|
[copia-like retrotransposon family, has a 5.4e-14 P-value blast match to gb|AAO73529.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
0.324 |
0.024 |
| 22 |
246238_at |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
0.312 |
-0.017 |
| 23 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
0.302 |
0.055 |
| 24 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
0.301 |
0.221 |
| 25 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.300 |
0.037 |
| 26 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
0.296 |
0.135 |
| 27 |
246733_at |
AT5G27660
|
DEG14, degradation of periplasmic proteins 14 |
0.293 |
0.068 |
| 28 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
0.292 |
0.013 |
| 29 |
250279_at |
AT5G13200
|
[GRAM domain family protein] |
0.283 |
0.045 |
| 30 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
0.283 |
-0.055 |
| 31 |
260783_at |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
0.271 |
0.373 |
| 32 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
0.271 |
-0.029 |
| 33 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.269 |
0.061 |
| 34 |
257373_at |
AT2G43140
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.267 |
-0.230 |
| 35 |
264271_at |
AT1G60270
|
BGLU6, beta glucosidase 6 |
0.267 |
0.033 |
| 36 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.267 |
0.209 |
| 37 |
250506_at |
AT5G09930
|
ABCF2, ATP-binding cassette F2 |
0.265 |
0.066 |
| 38 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
0.262 |
-0.082 |
| 39 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.262 |
0.034 |
| 40 |
254409_at |
AT4G21400
|
CRK28, cysteine-rich RLK (RECEPTOR-like protein kinase) 28 |
0.261 |
0.004 |
| 41 |
251221_at |
AT3G62550
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.260 |
0.023 |
| 42 |
250152_at |
AT5G15120
|
unknown |
0.258 |
-0.035 |
| 43 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
0.255 |
-0.010 |
| 44 |
252511_at |
AT3G46280
|
[protein kinase-related] |
0.250 |
0.020 |
| 45 |
251130_at |
AT5G01180
|
AtNPF8.2, ATPTR5, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 5, NPF8.2, NRT1/ PTR family 8.2, PTR5, peptide transporter 5 |
0.249 |
-0.031 |
| 46 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
0.246 |
-0.202 |
| 47 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
0.243 |
-0.116 |
| 48 |
258225_at |
AT3G15630
|
unknown |
0.243 |
-0.106 |
| 49 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
0.243 |
0.034 |
| 50 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.241 |
0.117 |
| 51 |
250127_at |
AT5G16380
|
unknown |
0.238 |
0.032 |
| 52 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
0.232 |
-0.232 |
| 53 |
254266_at |
AT4G23130
|
CRK5, cysteine-rich RLK (RECEPTOR-like protein kinase) 5, RLK6, RECEPTOR-LIKE PROTEIN KINASE 6 |
0.230 |
0.041 |
| 54 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
0.230 |
0.064 |
| 55 |
249777_at |
AT5G24210
|
[alpha/beta-Hydrolases superfamily protein] |
0.226 |
0.153 |
| 56 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
0.226 |
0.200 |
| 57 |
266983_at |
AT2G39400
|
[alpha/beta-Hydrolases superfamily protein] |
0.225 |
-0.025 |
| 58 |
256569_at |
AT3G19550
|
unknown |
0.224 |
-0.023 |
| 59 |
264767_at |
AT1G61380
|
SD1-29, S-domain-1 29 |
0.224 |
0.024 |
| 60 |
246791_at |
AT5G27280
|
[Zim17-type zinc finger protein] |
0.223 |
0.007 |
| 61 |
264756_at |
AT1G61370
|
[S-locus lectin protein kinase family protein] |
0.221 |
-0.100 |
| 62 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
0.220 |
-0.093 |
| 63 |
254301_at |
AT4G22790
|
[MATE efflux family protein] |
0.220 |
-0.046 |
| 64 |
250589_at |
AT5G07700
|
AtMYB76, myb domain protein 76, MYB76, myb domain protein 76 |
0.219 |
0.125 |
| 65 |
251054_at |
AT5G01540
|
LecRK-VI.2, L-type lectin receptor kinase-VI.2, LECRKA4.1, lectin receptor kinase a4.1 |
0.219 |
0.121 |
| 66 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
0.217 |
-0.107 |
| 67 |
258939_at |
AT3G10020
|
unknown |
0.217 |
-0.135 |
| 68 |
261947_at |
AT1G64470
|
[Ubiquitin-like superfamily protein] |
0.215 |
-0.191 |
| 69 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
0.212 |
0.037 |
| 70 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
0.210 |
0.013 |
| 71 |
255148_at |
AT4G08470
|
MAPKKK10, MEKK3, MAPK/ERK kinase kinase 3 |
0.210 |
0.153 |
| 72 |
246108_at |
AT5G28630
|
[glycine-rich protein] |
0.209 |
-0.117 |
| 73 |
254410_at |
AT4G21410
|
CRK29, cysteine-rich RLK (RECEPTOR-like protein kinase) 29 |
0.208 |
-0.082 |
| 74 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
0.208 |
0.019 |
| 75 |
247878_at |
AT5G57760
|
unknown |
0.207 |
-0.055 |
| 76 |
264741_at |
AT1G62290
|
[Saposin-like aspartyl protease family protein] |
0.206 |
0.040 |
| 77 |
262003_at |
AT1G64460
|
[Protein kinase superfamily protein] |
0.205 |
-0.096 |
| 78 |
248509_at |
AT5G50335
|
unknown |
0.204 |
-0.012 |
| 79 |
255607_at |
AT4G01130
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.203 |
-0.034 |
| 80 |
263391_at |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
0.201 |
0.136 |
| 81 |
258537_at |
AT3G04210
|
[Disease resistance protein (TIR-NBS class)] |
0.201 |
0.281 |
| 82 |
246817_at |
AT5G27240
|
[DNAJ heat shock N-terminal domain-containing protein] |
0.200 |
-0.042 |
| 83 |
266658_at |
AT2G25735
|
unknown |
0.195 |
0.141 |
| 84 |
256098_at |
AT1G13700
|
PGL1, 6-phosphogluconolactonase 1 |
0.195 |
0.031 |
| 85 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
0.195 |
0.259 |
| 86 |
263733_at |
AT1G60020
|
[copia-like retrotransposon family, has a 0. P-value blast match to GB:AAC02672 polyprotein (Ty1_Copia-element) (Arabidopsis arenosa)] |
0.195 |
0.002 |
| 87 |
246793_at |
AT5G27210
|
Cand8, candidate G-protein Coupled Receptor 8 |
0.192 |
-0.060 |
| 88 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
0.189 |
0.023 |
| 89 |
246417_at |
AT5G16990
|
[Zinc-binding dehydrogenase family protein] |
0.188 |
0.023 |
| 90 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
0.187 |
-0.014 |
| 91 |
259561_at |
AT1G21250
|
AtWAK1, PRO25, WAK1, cell wall-associated kinase 1 |
0.186 |
0.186 |
| 92 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
0.186 |
0.078 |
| 93 |
252387_at |
AT3G47800
|
[Galactose mutarotase-like superfamily protein] |
0.185 |
-0.021 |
| 94 |
245076_at |
AT2G23170
|
GH3.3 |
0.185 |
0.145 |
| 95 |
258379_at |
AT3G16700
|
[Fumarylacetoacetate (FAA) hydrolase family] |
0.184 |
0.009 |
| 96 |
246943_at |
AT5G25440
|
[Protein kinase superfamily protein] |
0.183 |
-0.044 |
| 97 |
259441_at |
AT1G02300
|
[Cysteine proteinases superfamily protein] |
0.183 |
0.009 |
| 98 |
256170_at |
AT1G51790
|
[Leucine-rich repeat protein kinase family protein] |
0.183 |
0.054 |
| 99 |
257805_at |
AT3G18830
|
ATPLT5, POLYOL TRANSPORTER 5, ATPMT5, ARABIDOPSIS THALIANA POLYOL/MONOSACCHARIDE TRANSPORTER 5, PMT5, polyol/monosaccharide transporter 5 |
0.182 |
0.048 |
| 100 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.181 |
0.070 |
| 101 |
245998_at |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
0.180 |
0.058 |
| 102 |
261254_at |
AT1G05805
|
AKS2, ABA-responsive kinase substrate 2 |
0.179 |
0.025 |
| 103 |
245449_at |
AT4G16870
|
[copia-like retrotransposon family, has a 0. P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
0.179 |
0.020 |
| 104 |
256455_at |
AT1G75190
|
unknown |
0.178 |
-0.069 |
| 105 |
258923_at |
AT3G10450
|
SCPL7, serine carboxypeptidase-like 7 |
0.177 |
-0.085 |
| 106 |
265713_at |
AT2G03530
|
ATUPS2, ARABIDOPSIS THALIANA UREIDE PERMEASE 2, UPS2, ureide permease 2 |
0.176 |
-0.050 |
| 107 |
246683_at |
AT5G33300
|
[chromosome-associated kinesin-related] |
0.173 |
-0.052 |
| 108 |
244943_at |
ATMG00070
|
NAD9, NADH dehydrogenase subunit 9 |
0.173 |
-0.102 |
| 109 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
0.172 |
0.074 |
| 110 |
255913_at |
AT1G66980
|
GDPDL2, Glycerophosphodiester phosphodiesterase (GDPD) like 2, SNC4, suppressor of npr1-1 constitutive 4 |
0.172 |
-0.173 |
| 111 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
0.171 |
0.021 |
| 112 |
249948_at |
AT5G19210
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.170 |
-0.037 |
| 113 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
0.170 |
-0.118 |
| 114 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
0.170 |
-0.118 |
| 115 |
252820_at |
AT3G42640
|
AHA8, H(+)-ATPase 8, HA8, H(+)-ATPase 8 |
0.170 |
0.045 |
| 116 |
260656_at |
AT1G19380
|
unknown |
0.169 |
0.111 |
| 117 |
263126_at |
AT1G78460
|
[SOUL heme-binding family protein] |
0.168 |
0.041 |
| 118 |
254242_at |
AT4G23200
|
CRK12, cysteine-rich RLK (RECEPTOR-like protein kinase) 12 |
0.167 |
0.058 |
| 119 |
254093_at |
AT4G25110
|
AtMC2, metacaspase 2, AtMCP1c, metacaspase 1c, MC2, metacaspase 2, MCP1c, metacaspase 1c |
0.166 |
0.134 |
| 120 |
260799_at |
AT1G78270
|
AtUGT85A4, UDP-glucosyl transferase 85A4, UGT85A4, UDP-glucosyl transferase 85A4 |
0.165 |
0.006 |
| 121 |
249919_at |
AT5G19250
|
[Glycoprotein membrane precursor GPI-anchored] |
0.165 |
-0.006 |
| 122 |
250455_at |
AT5G09980
|
PROPEP4, elicitor peptide 4 precursor |
0.165 |
0.005 |
| 123 |
250548_at |
AT5G08120
|
MPB2C, movement protein binding protein 2C |
0.164 |
-0.089 |
| 124 |
259560_at |
AT1G21270
|
WAK2, wall-associated kinase 2 |
0.164 |
-0.023 |
| 125 |
252464_at |
AT3G47160
|
[RING/U-box superfamily protein] |
0.163 |
-0.108 |
| 126 |
267081_at |
AT2G41210
|
PIP5K5, phosphatidylinositol- 4-phosphate 5-kinase 5 |
0.163 |
-0.055 |
| 127 |
251919_at |
AT3G53800
|
Fes1B, Fes1B |
0.163 |
0.083 |
| 128 |
267230_at |
AT2G44080
|
ARL, ARGOS-like |
0.163 |
-0.007 |
| 129 |
264510_at |
AT1G09530
|
PAP3, PHYTOCHROME-ASSOCIATED PROTEIN 3, PIF3, phytochrome interacting factor 3, POC1, PHOTOCURRENT 1 |
0.161 |
-0.009 |
| 130 |
266500_at |
AT2G06925
|
ATSPLA2-ALPHA, PHOSPHOLIPASE A2-ALPHA, PLA2-ALPHA |
0.160 |
-0.020 |
| 131 |
256664_at |
AT3G12040
|
[DNA-3-methyladenine glycosylase (MAG)] |
0.160 |
0.020 |
| 132 |
246082_at |
AT5G20480
|
EFR, EF-TU receptor |
0.160 |
0.065 |
| 133 |
249941_at |
AT5G22270
|
unknown |
0.159 |
-0.005 |
| 134 |
266016_at |
AT2G18670
|
[RING/U-box superfamily protein] |
0.159 |
-0.056 |
| 135 |
250132_at |
AT5G16560
|
KAN, KANADI, KAN1, KANADI 1 |
0.158 |
-0.103 |
| 136 |
265246_at |
AT2G43050
|
ATPMEPCRD |
0.157 |
-0.321 |
| 137 |
257677_at |
AT3G20290
|
ATEHD1, EPS15 homology domain 1, EHD1, EPS15 homology domain 1 |
0.157 |
-0.060 |
| 138 |
261763_at |
AT1G15520
|
ABCG40, ATP-binding cassette G40, ATABCG40, Arabidopsis thaliana ATP-binding cassette G40, ATPDR12, PLEIOTROPIC DRUG RESISTANCE 12, PDR12, pleiotropic drug resistance 12 |
0.157 |
0.089 |
| 139 |
256852_at |
AT3G18610
|
ATNUC-L2, nucleolin like 2, NUC-L2, nucleolin like 2, PARLL1, PARALLEL1-LIKE 1 |
0.156 |
0.084 |
| 140 |
257929_at |
AT3G16980
|
NRPB9A, NRPD9A, NRPE9A |
0.154 |
0.026 |
| 141 |
253122_at |
AT4G35987
|
CaM KMT, calmodulin N-methyltransferase |
0.154 |
-0.090 |
| 142 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
0.154 |
-0.019 |
| 143 |
264916_at |
AT1G60810
|
ACLA-2, ATP-citrate lyase A-2 |
0.153 |
-0.037 |
| 144 |
262309_at |
AT1G70820
|
[phosphoglucomutase, putative / glucose phosphomutase, putative] |
0.150 |
-0.050 |
| 145 |
260671_at |
AT1G19310
|
[RING/U-box superfamily protein] |
0.150 |
-0.039 |
| 146 |
257113_at |
AT3G20130
|
CYP705A22, cytochrome P450, family 705, subfamily A, polypeptide 22, GPS1, gravity persistence signal 1 |
0.149 |
0.066 |
| 147 |
259399_at |
AT1G17710
|
AtPEPC1, Arabidopsis thaliana phosphoethanolamine/phosphocholine phosphatase 1, PEPC1, phosphoethanolamine/phosphocholine phosphatase 1 |
0.148 |
0.243 |
| 148 |
254926_at |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
-0.945 |
0.054 |
| 149 |
259384_at |
AT3G16450
|
JAL33, Jacalin-related lectin 33 |
-0.920 |
-0.068 |
| 150 |
246825_at |
AT5G26260
|
[TRAF-like family protein] |
-0.878 |
-0.062 |
| 151 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.782 |
-0.331 |
| 152 |
257057_at |
AT3G15310
|
unknown |
-0.774 |
-0.037 |
| 153 |
250063_at |
AT5G17880
|
CSA1, constitutive shade-avoidance1 |
-0.770 |
-0.043 |
| 154 |
246888_at |
AT5G26270
|
unknown |
-0.699 |
-0.052 |
| 155 |
250433_at |
AT5G10400
|
H3.1, histone 3.1 |
-0.698 |
-0.014 |
| 156 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
-0.587 |
-0.021 |
| 157 |
250064_at |
AT5G17890
|
CHS3, CHILLING SENSITIVE 3, DAR4, DA1-related protein 4 |
-0.580 |
-0.010 |
| 158 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.580 |
-0.118 |
| 159 |
249774_at |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
-0.564 |
0.280 |
| 160 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
-0.536 |
0.029 |
| 161 |
246478_at |
AT5G15980
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.517 |
0.007 |
| 162 |
245982_at |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
-0.492 |
0.081 |
| 163 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
-0.476 |
0.019 |
| 164 |
249476_at |
AT5G38910
|
[RmlC-like cupins superfamily protein] |
-0.455 |
-0.051 |
| 165 |
249724_at |
AT5G35450
|
[Disease resistance protein (CC-NBS-LRR class) family] |
-0.435 |
-0.176 |
| 166 |
250528_at |
AT5G08600
|
[U3 ribonucleoprotein (Utp) family protein] |
-0.410 |
0.054 |
| 167 |
254201_at |
AT4G24130
|
unknown |
-0.396 |
0.144 |
| 168 |
246002_at |
AT5G20740
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.391 |
0.028 |
| 169 |
253514_at |
AT4G31805
|
POLAR, POLAR LOCALIZATION DURING ASYMMETRIC DIVISION AND REDISTRIBUTION |
-0.384 |
-0.132 |
| 170 |
246542_at |
AT5G15020
|
SNL2, SIN3-like 2 |
-0.381 |
0.001 |
| 171 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-0.381 |
0.168 |
| 172 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.376 |
0.077 |
| 173 |
250899_at |
AT5G03340
|
AtCDC48C, cell division cycle 48C |
-0.362 |
0.010 |
| 174 |
250696_at |
AT5G06790
|
unknown |
-0.359 |
0.138 |
| 175 |
246001_at |
AT5G20790
|
unknown |
-0.357 |
0.052 |
| 176 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
-0.357 |
-0.082 |
| 177 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
-0.350 |
0.074 |
| 178 |
266141_at |
AT2G02120
|
LCR70, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 70, PDF2.1 |
-0.349 |
0.164 |
| 179 |
246818_at |
AT5G27270
|
EMB976, EMBRYO DEFECTIVE 976 |
-0.348 |
-0.010 |
| 180 |
257112_at |
AT3G20120
|
CYP705A21, cytochrome P450, family 705, subfamily A, polypeptide 21 |
-0.347 |
-0.077 |
| 181 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
-0.346 |
-0.056 |
| 182 |
259573_at |
AT1G20390
|
[gypsy-like retrotransposon family, has a 1.5e-251 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
-0.344 |
-0.196 |
| 183 |
246497_at |
AT5G16220
|
[Octicosapeptide/Phox/Bem1p family protein] |
-0.341 |
0.063 |
| 184 |
249675_at |
AT5G35940
|
[Mannose-binding lectin superfamily protein] |
-0.339 |
-0.070 |
| 185 |
249727_at |
AT5G35490
|
ATMRU1, ARABIDOPSIS MTO 1 RESPONDING UP 1, MRU1, MTO 1 RESPONDING UP 1 |
-0.334 |
-0.106 |
| 186 |
249860_at |
AT5G22860
|
[Serine carboxypeptidase S28 family protein] |
-0.326 |
-0.115 |
| 187 |
256320_at |
AT3G12170
|
[Chaperone DnaJ-domain superfamily protein] |
-0.325 |
-0.047 |
| 188 |
254371_at |
AT4G21760
|
BGLU47, beta-glucosidase 47 |
-0.324 |
0.080 |
| 189 |
246966_at |
AT5G24850
|
CRY3, cryptochrome 3 |
-0.324 |
-0.007 |
| 190 |
258064_at |
AT3G14680
|
CYP72A14, cytochrome P450, family 72, subfamily A, polypeptide 14 |
-0.320 |
0.178 |
| 191 |
251024_at |
AT5G02180
|
[Transmembrane amino acid transporter family protein] |
-0.316 |
0.043 |
| 192 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
-0.312 |
-0.227 |
| 193 |
254550_at |
AT4G19690
|
ATIRT1, ARABIDOPSIS IRON-REGULATED TRANSPORTER 1, IRT1, iron-regulated transporter 1 |
-0.305 |
0.410 |
| 194 |
254251_at |
AT4G23300
|
CRK22, cysteine-rich RLK (RECEPTOR-like protein kinase) 22 |
-0.305 |
0.147 |
| 195 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
-0.296 |
0.191 |
| 196 |
251050_at |
AT5G02440
|
unknown |
-0.296 |
-0.190 |
| 197 |
254313_at |
AT4G22460
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.295 |
0.052 |
| 198 |
250382_at |
AT5G11580
|
[Regulator of chromosome condensation (RCC1) family protein] |
-0.295 |
-0.033 |
| 199 |
246122_at |
AT5G20380
|
PHT4;5, phosphate transporter 4;5 |
-0.289 |
0.006 |
| 200 |
250771_at |
AT5G05400
|
[LRR and NB-ARC domains-containing disease resistance protein] |
-0.288 |
-0.146 |
| 201 |
256257_at |
AT3G11240
|
ATATE2, ATE2, arginine-tRNA protein transferase 2 |
-0.286 |
-0.033 |
| 202 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
-0.281 |
0.150 |
| 203 |
254362_at |
AT4G22160
|
unknown |
-0.280 |
0.005 |
| 204 |
254078_at |
AT4G25710
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.280 |
-0.004 |
| 205 |
260582_at |
AT2G47200
|
unknown |
-0.274 |
-0.012 |
| 206 |
251093_at |
AT5G01360
|
TBL3, TRICHOME BIREFRINGENCE-LIKE 3 |
-0.272 |
0.021 |
| 207 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
-0.268 |
0.086 |
| 208 |
254437_s_at |
AT3G06433
|
[pseudogene] |
-0.267 |
0.004 |
| 209 |
264166_at |
AT1G65370
|
[TRAF-like family protein] |
-0.266 |
-0.001 |
| 210 |
245965_at |
AT5G19730
|
[Pectin lyase-like superfamily protein] |
-0.266 |
0.059 |
| 211 |
249813_at |
AT5G23940
|
DCR, DEFECTIVE IN CUTICULAR RIDGES, EMB3009, EMBRYO DEFECTIVE 3009, PEL3, PERMEABLE LEAVES3 |
-0.263 |
0.043 |
| 212 |
257235_at |
AT3G15060
|
AtRABA1g, RAB GTPase homolog A1G, RABA1g, RAB GTPase homolog A1G |
-0.261 |
0.057 |
| 213 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.258 |
0.247 |
| 214 |
250537_at |
AT5G08565
|
[Transcription initiation Spt4-like protein] |
-0.256 |
0.113 |
| 215 |
257291_at |
AT3G15590
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.255 |
0.015 |
| 216 |
258113_at |
AT3G14650
|
CYP72A11, cytochrome P450, family 72, subfamily A, polypeptide 11 |
-0.254 |
-0.029 |
| 217 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
-0.253 |
0.063 |
| 218 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
-0.250 |
0.280 |
| 219 |
250859_at |
AT5G04660
|
CYP77A4, cytochrome P450, family 77, subfamily A, polypeptide 4 |
-0.249 |
0.063 |
| 220 |
250084_at |
AT5G17240
|
SDG40, SET domain group 40 |
-0.246 |
0.105 |
| 221 |
256619_at |
AT3G24460
|
[Serinc-domain containing serine and sphingolipid biosynthesis protein] |
-0.243 |
-0.085 |
| 222 |
249832_at |
AT5G23400
|
[Leucine-rich repeat (LRR) family protein] |
-0.241 |
-0.108 |
| 223 |
249622_at |
AT5G37550
|
unknown |
-0.233 |
0.059 |
| 224 |
250646_at |
AT5G06720
|
ATPA2, peroxidase 2, AtPRX53, PA2, peroxidase 2, PRX53, peroxidase 53 |
-0.233 |
0.004 |
| 225 |
256774_at |
AT3G13760
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.232 |
-0.130 |
| 226 |
257978_at |
AT3G20860
|
ATNEK5, NIMA-related kinase 5, NEK5, NIMA-related kinase 5 |
-0.230 |
-0.046 |
| 227 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
-0.230 |
0.098 |
| 228 |
261545_at |
AT1G63530
|
unknown |
-0.228 |
0.470 |
| 229 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
-0.224 |
0.285 |
| 230 |
254185_at |
AT4G23990
|
ATCSLG3, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE-LIKE G3, CSLG3, cellulose synthase like G3 |
-0.224 |
-0.041 |
| 231 |
249750_at |
AT5G24570
|
unknown |
-0.223 |
0.028 |
| 232 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
-0.222 |
0.564 |
| 233 |
247055_at |
AT5G66740
|
unknown |
-0.218 |
0.098 |
| 234 |
254340_at |
AT4G22120
|
[ERD (early-responsive to dehydration stress) family protein] |
-0.217 |
-0.008 |
| 235 |
263005_at |
AT1G54540
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.217 |
0.155 |
| 236 |
250098_at |
AT5G17350
|
unknown |
-0.215 |
-0.024 |
| 237 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
-0.213 |
0.055 |
| 238 |
259209_at |
AT3G09160
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.213 |
0.004 |
| 239 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.213 |
0.122 |
| 240 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
-0.212 |
0.129 |
| 241 |
249101_at |
AT5G43580
|
UPI, UNUSUAL SERINE PROTEASE INHIBITOR |
-0.211 |
-0.184 |
| 242 |
256935_at |
AT3G22570
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.210 |
-0.115 |
| 243 |
250598_at |
AT5G07690
|
ATMYB29, myb domain protein 29, MYB29, myb domain protein 29, PMG2, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 2 |
-0.210 |
0.064 |
| 244 |
259629_at |
AT1G56510
|
ADR2, ACTIVATED DISEASE RESISTANCE 2, WRR4, WHITE RUST RESISTANCE 4 |
-0.209 |
0.020 |
| 245 |
252958_at |
AT4G38620
|
ATMYB4, myb domain protein 4, MYB4, myb domain protein 4 |
-0.207 |
0.050 |
| 246 |
249759_at |
AT5G24380
|
ATYSL2, YELLOW STRIPE LIKE 2, YSL2, YELLOW STRIPE like 2 |
-0.206 |
0.072 |
| 247 |
263007_at |
AT1G54260
|
[winged-helix DNA-binding transcription factor family protein] |
-0.205 |
0.049 |
| 248 |
256527_at |
AT1G66100
|
[Plant thionin] |
-0.205 |
-0.058 |
| 249 |
250050_at |
AT5G17790
|
VAR3, VARIEGATED 3 |
-0.204 |
0.029 |
| 250 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
-0.203 |
0.065 |
| 251 |
258807_at |
AT3G04030
|
MYR2 |
-0.203 |
0.032 |
| 252 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.200 |
-0.091 |
| 253 |
246494_at |
AT5G16190
|
ATCSLA11, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE A11, CSLA11, cellulose synthase like A11 |
-0.200 |
0.079 |
| 254 |
253185_at |
AT4G35240
|
unknown |
-0.198 |
-0.081 |
| 255 |
248048_at |
AT5G56080
|
ATNAS2, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 2, NAS2, nicotianamine synthase 2 |
-0.197 |
0.307 |
| 256 |
246735_at |
AT5G27670
|
HTA7, histone H2A 7 |
-0.197 |
0.002 |
| 257 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
-0.197 |
0.139 |
| 258 |
256275_at |
AT3G12110
|
ACT11, actin-11 |
-0.196 |
-0.005 |
| 259 |
249882_at |
AT5G22890
|
STOP2, sensitive to proton rhizotoxicity 2 |
-0.195 |
0.081 |
| 260 |
248246_at |
AT5G53200
|
TRY, TRIPTYCHON |
-0.195 |
-0.043 |
| 261 |
261514_at |
AT1G71870
|
[MATE efflux family protein] |
-0.192 |
0.095 |
| 262 |
259445_at |
AT1G02400
|
ATGA2OX4, Arabidopsis thaliana gibberellin 2-oxidase 4, ATGA2OX6, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 6, DTA1, DOWNSTREAM TARGET OF AGL15 1, GA2OX6, gibberellin 2-oxidase 6 |
-0.192 |
-0.064 |
| 263 |
249686_at |
AT5G36140
|
CYP716A2, cytochrome P450, family 716, subfamily A, polypeptide 2 |
-0.191 |
0.079 |
| 264 |
251020_at |
AT5G02270
|
ABCI20, ATP-binding cassette I20, ATNAP9, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN 9, NAP9, non-intrinsic ABC protein 9 |
-0.191 |
0.023 |
| 265 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
-0.190 |
0.117 |
| 266 |
254352_at |
AT4G22320
|
unknown |
-0.190 |
0.021 |
| 267 |
248978_at |
AT5G45070
|
AtPP2-A8, phloem protein 2-A8, PP2-A8, phloem protein 2-A8 |
-0.188 |
0.319 |
| 268 |
249205_at |
AT5G42600
|
MRN1, marneral synthase 1 |
-0.188 |
-0.229 |
| 269 |
249898_at |
AT5G22510
|
A/N-InvE, alkaline/neutral invertase E, At-A/N-InvE, Arabidopsis alkaline/neutral invertase E, INV-E, alkaline/neutral invertase E |
-0.187 |
-0.015 |
| 270 |
249934_at |
AT5G22410
|
RHS18, root hair specific 18 |
-0.187 |
-0.352 |
| 271 |
251545_at |
AT3G58810
|
ATMTP3, ARABIDOPSIS METAL TOLERANCE PROTEIN 3, ATMTPA2, MTP3, METAL TOLERANCE PROTEIN 3, MTPA2, metal tolerance protein A2 |
-0.186 |
0.248 |
| 272 |
251293_at |
AT3G61930
|
unknown |
-0.184 |
0.262 |
| 273 |
262212_at |
AT1G74890
|
ARR15, response regulator 15 |
-0.182 |
-0.015 |
| 274 |
246395_at |
AT1G58170
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.182 |
0.121 |
| 275 |
246120_at |
AT5G20360
|
Phox3, Phox3 |
-0.180 |
0.048 |
| 276 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
-0.180 |
0.052 |
| 277 |
259142_at |
AT3G10200
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.179 |
0.072 |
| 278 |
249124_at |
AT5G43740
|
[Disease resistance protein (CC-NBS-LRR class) family] |
-0.177 |
-0.091 |
| 279 |
266516_at |
AT2G47880
|
[Glutaredoxin family protein] |
-0.177 |
0.074 |
| 280 |
258201_at |
AT3G13910
|
unknown |
-0.176 |
0.011 |
| 281 |
251107_at |
AT5G01610
|
unknown |
-0.175 |
-0.250 |
| 282 |
246505_at |
AT5G16250
|
unknown |
-0.175 |
-0.015 |
| 283 |
249639_at |
AT5G36930
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.174 |
0.008 |
| 284 |
246702_at |
AT5G28050
|
GSDA, guanosine deaminase |
-0.174 |
-0.009 |
| 285 |
261150_at |
AT1G19640
|
JMT, jasmonic acid carboxyl methyltransferase |
-0.174 |
0.052 |
| 286 |
249874_at |
AT5G23070
|
AtTK1b, TK1b, thymidine kinase 1b |
-0.172 |
0.007 |
| 287 |
250616_at |
AT5G07280
|
EMS1, EXCESS MICROSPOROCYTES1, EXS, EXTRA SPOROGENOUS CELLS |
-0.171 |
0.035 |
| 288 |
246882_at |
AT5G26180
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.171 |
-0.044 |
| 289 |
246992_at |
AT5G67430
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.170 |
0.011 |
| 290 |
257009_at |
AT3G14160
|
[2-oxoglutarate-dependent dioxygenase family protein] |
-0.170 |
0.029 |
| 291 |
256021_at |
AT1G58270
|
ZW9 |
-0.170 |
-0.059 |
| 292 |
257066_at |
AT3G18280
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.169 |
0.064 |
| 293 |
250265_at |
AT5G12900
|
unknown |
-0.169 |
-0.056 |