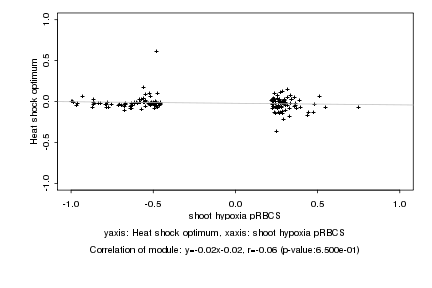
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
shoot hypoxia pRBCS |
Log2 signal ratio
Heat shock optimum |
| 1 |
245385_at |
AT4G14020
|
[Rapid alkalinization factor (RALF) family protein] |
0.748 |
-0.066 |
| 2 |
260672_at |
AT1G19480
|
[DNA glycosylase superfamily protein] |
0.543 |
-0.064 |
| 3 |
248102_at |
AT5G55140
|
[ribosomal protein L30 family protein] |
0.507 |
0.062 |
| 4 |
246518_at |
AT5G15770
|
AtGNA1, glucose-6-phosphate acetyltransferase 1, GNA1, glucose-6-phosphate acetyltransferase 1 |
0.475 |
-0.029 |
| 5 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
0.470 |
-0.124 |
| 6 |
259432_at |
AT1G01520
|
ASG4, ALTERED SEED GERMINATION 4 |
0.439 |
-0.126 |
| 7 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
0.434 |
-0.159 |
| 8 |
264201_at |
AT1G22630
|
unknown |
0.395 |
-0.068 |
| 9 |
251985_at |
AT3G53220
|
[Thioredoxin superfamily protein] |
0.388 |
0.017 |
| 10 |
249422_at |
AT5G39760
|
AtHB23, homeobox protein 23, HB23, homeobox protein 23, ZHD10, ZINC FINGER HOMEODOMAIN 10 |
0.369 |
-0.081 |
| 11 |
266717_at |
AT2G46735
|
unknown |
0.365 |
-0.019 |
| 12 |
261629_at |
AT1G49980
|
[DNA/RNA polymerases superfamily protein] |
0.360 |
-0.047 |
| 13 |
261110_at |
AT1G75440
|
UBC16, ubiquitin-conjugating enzyme 16 |
0.356 |
0.057 |
| 14 |
247252_at |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
0.354 |
-0.052 |
| 15 |
253607_at |
AT4G30330
|
[Small nuclear ribonucleoprotein family protein] |
0.339 |
0.028 |
| 16 |
249441_at |
AT5G39730
|
[AIG2-like (avirulence induced gene) family protein] |
0.336 |
0.026 |
| 17 |
253726_at |
AT4G29430
|
rps15ae, ribosomal protein S15A E |
0.330 |
0.077 |
| 18 |
259423_at |
AT1G13880
|
[ELM2 domain-containing protein] |
0.329 |
-0.017 |
| 19 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
0.325 |
-0.172 |
| 20 |
261516_at |
AT1G71750
|
HGPT, Hypoxanthine-guanine phosphoribosyltransferase |
0.324 |
-0.081 |
| 21 |
262314_at |
AT1G70810
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.315 |
0.058 |
| 22 |
266237_at |
AT2G29540
|
ATRPAC14, ATRPC14, RNApolymerase 14 kDa subunit, RPC14, RNApolymerase 14 kDa subunit |
0.313 |
0.151 |
| 23 |
249648_at |
AT5G37050
|
unknown |
0.313 |
-0.042 |
| 24 |
246757_at |
AT5G27940
|
WPP3, WPP domain protein 3 |
0.304 |
-0.005 |
| 25 |
260776_at |
AT1G14580
|
[C2H2-like zinc finger protein] |
0.303 |
-0.047 |
| 26 |
257081_at |
AT3G30460
|
[RING/U-box superfamily protein] |
0.302 |
-0.107 |
| 27 |
249834_at |
AT5G23440
|
FTRA1, ferredoxin/thioredoxin reductase subunit A (variable subunit) 1 |
0.300 |
-0.012 |
| 28 |
247632_at |
AT5G60460
|
[Preprotein translocase Sec, Sec61-beta subunit protein] |
0.296 |
0.027 |
| 29 |
265044_at |
AT1G03950
|
VPS2.3, vacuolar protein sorting-associated protein 2.3 |
0.296 |
-0.036 |
| 30 |
257058_at |
AT3G15352
|
ATCOX17, ARABIDOPSIS THALIANA CYTOCHROME C OXIDASE 17, COX17, cytochrome c oxidase 17 |
0.295 |
0.013 |
| 31 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
0.289 |
-0.214 |
| 32 |
253917_at |
AT4G27380
|
unknown |
0.288 |
-0.008 |
| 33 |
253830_at |
AT4G27652
|
unknown |
0.286 |
0.133 |
| 34 |
250881_at |
AT5G04080
|
unknown |
0.286 |
-0.118 |
| 35 |
252661_at |
AT3G44450
|
unknown |
0.283 |
-0.137 |
| 36 |
256653_at |
AT3G18870
|
[Mitochondrial transcription termination factor family protein] |
0.283 |
0.003 |
| 37 |
262049_at |
AT1G80180
|
unknown |
0.281 |
-0.012 |
| 38 |
248028_at |
AT5G55620
|
unknown |
0.278 |
-0.057 |
| 39 |
249260_at |
AT5G41680
|
[Protein kinase superfamily protein] |
0.277 |
-0.073 |
| 40 |
267636_at |
AT2G42110
|
unknown |
0.273 |
0.114 |
| 41 |
266566_at |
AT2G24040
|
[Low temperature and salt responsive protein family] |
0.273 |
-0.006 |
| 42 |
265183_at |
AT1G23750
|
[Nucleic acid-binding, OB-fold-like protein] |
0.272 |
0.012 |
| 43 |
253305_at |
AT4G33666
|
unknown |
0.271 |
-0.018 |
| 44 |
263597_at |
AT2G01870
|
unknown |
0.269 |
-0.008 |
| 45 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.269 |
-0.126 |
| 46 |
249878_at |
AT5G23090
|
NF-YB13, nuclear factor Y, subunit B13 |
0.266 |
0.019 |
| 47 |
258595_at |
AT3G04500
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.265 |
0.030 |
| 48 |
246462_at |
AT5G16940
|
[carbon-sulfur lyases] |
0.265 |
-0.139 |
| 49 |
245747_at |
AT1G51100
|
CRR41, CHLORORESPIRATORY REDUCTION 41 |
0.264 |
0.031 |
| 50 |
247793_at |
AT5G58650
|
PSY1, plant peptide containing sulfated tyrosine 1 |
0.264 |
-0.072 |
| 51 |
246653_at |
AT5G35200
|
[ENTH/ANTH/VHS superfamily protein] |
0.260 |
0.009 |
| 52 |
249109_at |
AT5G43700
|
ATAUX2-11, AUXIN INDUCIBLE 2-11, IAA4, indole-3-acetic acid inducible 4 |
0.260 |
-0.041 |
| 53 |
248261_at |
AT5G53280
|
PDV1, PLASTID DIVISION1 |
0.259 |
-0.132 |
| 54 |
254624_at |
AT4G18580
|
unknown |
0.254 |
0.082 |
| 55 |
261247_at |
AT1G20070
|
unknown |
0.253 |
-0.078 |
| 56 |
258002_at |
AT3G28930
|
AIG2, AVRRPT2-INDUCED GENE 2 |
0.250 |
0.043 |
| 57 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
0.249 |
-0.365 |
| 58 |
254611_at |
AT4G19095
|
unknown |
0.249 |
-0.055 |
| 59 |
245367_at |
AT4G16265
|
NRPB9B, NRPD9B, NRPE9B |
0.243 |
-0.013 |
| 60 |
246936_at |
AT5G25360
|
unknown |
0.242 |
-0.063 |
| 61 |
264692_at |
AT1G70000
|
[myb-like transcription factor family protein] |
0.241 |
-0.141 |
| 62 |
252041_at |
AT3G52090
|
ATRPB13.6, NRPB11, NRPD11, NRPE11 |
0.239 |
0.005 |
| 63 |
252565_at |
AT3G46000
|
ADF2, actin depolymerizing factor 2 |
0.237 |
0.034 |
| 64 |
251790_at |
AT3G55470
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.236 |
0.034 |
| 65 |
251036_at |
AT5G02160
|
unknown |
0.235 |
0.036 |
| 66 |
265025_at |
AT1G24575
|
unknown |
0.232 |
0.105 |
| 67 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
0.232 |
-0.126 |
| 68 |
262367_at |
AT1G73030
|
CHMP1A, CHARGED MULTIVESICULAR BODY PROTEIN/CHROMATIN MODIFYING PROTEIN1A, VPS46.2 |
0.231 |
0.043 |
| 69 |
252935_at |
AT4G39150
|
[DNAJ heat shock N-terminal domain-containing protein] |
0.229 |
-0.056 |
| 70 |
262389_at |
AT1G49270
|
AtPERK7, proline-rich extensin-like receptor kinase 7, PERK7, proline-rich extensin-like receptor kinase 7 |
0.228 |
-0.022 |
| 71 |
261409_at |
AT1G07640
|
OBP2, URP3, UAS-TAGGED ROOT PATTERNING3 |
0.228 |
-0.055 |
| 72 |
260004_at |
AT1G67860
|
unknown |
0.227 |
-0.051 |
| 73 |
263630_at |
AT2G04845
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.225 |
-0.034 |
| 74 |
246007_at |
AT5G08410
|
FTRA2, ferredoxin/thioredoxin reductase subunit A (variable subunit) 2 |
0.224 |
0.021 |
| 75 |
265676_at |
AT2G32070
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
0.223 |
0.034 |
| 76 |
246497_at |
AT5G16220
|
[Octicosapeptide/Phox/Bem1p family protein] |
0.222 |
0.007 |
| 77 |
249863_at |
AT5G22950
|
VPS24.1 |
0.222 |
-0.012 |
| 78 |
256527_at |
AT1G66100
|
[Plant thionin] |
0.220 |
-0.083 |
| 79 |
251393_at |
AT3G60640
|
ATG8G, AUTOPHAGY 8G |
0.220 |
0.032 |
| 80 |
254355_at |
AT4G22380
|
[Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein] |
0.218 |
0.016 |
| 81 |
244981_at |
ATCG00770
|
RPS8, ribosomal protein S8 |
-1.002 |
0.001 |
| 82 |
266393_at |
AT2G41260
|
ATM17, M17 |
-0.996 |
0.011 |
| 83 |
244985_at |
ATCG00810
|
RPL22, ribosomal protein L22 |
-0.991 |
-0.009 |
| 84 |
245025_at |
ATCG00130
|
ATPF |
-0.972 |
-0.037 |
| 85 |
265211_at |
AT2G36640
|
ATECP63, embryonic cell protein 63, ECP63, embryonic cell protein 63 |
-0.962 |
-0.017 |
| 86 |
244966_at |
ATCG00600
|
PETG |
-0.933 |
0.062 |
| 87 |
245049_at |
ATCG00050
|
RPS16, ribosomal protein S16 |
-0.873 |
-0.066 |
| 88 |
244973_at |
ATCG00690
|
PSBT, photosystem II reaction center protein T, PSBTC |
-0.867 |
-0.026 |
| 89 |
266353_at |
AT2G01520
|
MLP328, MLP-like protein 328, ZCE1, (Zusammen-CA)-enhanced 1 |
-0.865 |
0.032 |
| 90 |
244980_at |
ATCG00760
|
RPL36, ribosomal protein L36 |
-0.864 |
-0.009 |
| 91 |
244970_at |
ATCG00660
|
RPL20, ribosomal protein L20 |
-0.858 |
-0.014 |
| 92 |
244965_at |
ATCG00590
|
ORF31 |
-0.838 |
-0.018 |
| 93 |
244936_at |
ATCG01100
|
NDHA |
-0.826 |
-0.016 |
| 94 |
266544_at |
AT2G35300
|
AtLEA4-2, late embryogenesis abundant 4-2, LEA18, late embryogenesis abundant 18, LEA4-2, late embryogenesis abundant 4-2 |
-0.792 |
-0.026 |
| 95 |
257130_at |
AT3G20210
|
DELTA-VPE, delta vacuolar processing enzyme, DELTAVPE, DELTA VACUOLAR PROCESSING ENZYME |
-0.786 |
-0.027 |
| 96 |
257334_at |
ATMG01370
|
ORF111D |
-0.786 |
-0.068 |
| 97 |
244983_at |
ATCG00790
|
RPL16, ribosomal protein L16 |
-0.782 |
-0.007 |
| 98 |
245016_at |
ATCG00500
|
ACCD, acetyl-CoA carboxylase carboxyl transferase subunit beta |
-0.777 |
-0.063 |
| 99 |
245008_at |
ATCG00360
|
YCF3 |
-0.759 |
-0.027 |
| 100 |
244932_at |
ATCG01060
|
PSAC |
-0.716 |
-0.047 |
| 101 |
245017_at |
ATCG00510
|
PSAI, photsystem I subunit I |
-0.710 |
-0.025 |
| 102 |
258347_at |
AT3G17520
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.696 |
-0.045 |
| 103 |
244982_at |
ATCG00780
|
RPL14, ribosomal protein L14 |
-0.677 |
-0.026 |
| 104 |
245002_at |
ATCG00270
|
PSBD, photosystem II reaction center protein D |
-0.676 |
-0.104 |
| 105 |
251838_at |
AT3G54940
|
[Papain family cysteine protease] |
-0.676 |
-0.054 |
| 106 |
252019_at |
AT3G53040
|
[late embryogenesis abundant protein, putative / LEA protein, putative] |
-0.675 |
-0.014 |
| 107 |
244933_at |
ATCG01070
|
NDHE |
-0.671 |
-0.025 |
| 108 |
245011_at |
ATCG00430
|
PSBG, photosystem II reaction center protein G |
-0.643 |
-0.080 |
| 109 |
261981_at |
AT1G33811
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.643 |
-0.060 |
| 110 |
244998_at |
ATCG00180
|
RPOC1 |
-0.636 |
-0.037 |
| 111 |
245003_at |
ATCG00280
|
PSBC, photosystem II reaction center protein C |
-0.634 |
-0.078 |
| 112 |
256814_at |
AT3G21370
|
BGLU19, beta glucosidase 19 |
-0.633 |
-0.022 |
| 113 |
244934_at |
ATCG01080
|
NDHG |
-0.628 |
-0.047 |
| 114 |
245026_at |
ATCG00140
|
ATPH |
-0.627 |
-0.046 |
| 115 |
244996_at |
ATCG00160
|
RPS2, ribosomal protein S2 |
-0.617 |
-0.022 |
| 116 |
244967_at |
ATCG00630
|
PSAJ |
-0.617 |
-0.007 |
| 117 |
244963_at |
ATCG00570
|
PSBF, photosystem II reaction center protein F |
-0.604 |
-0.006 |
| 118 |
244975_at |
ATCG00710
|
PSBH, photosystem II reaction center protein H |
-0.600 |
-0.024 |
| 119 |
245021_at |
ATCG00550
|
PSBJ, photosystem II reaction center protein J |
-0.586 |
-0.004 |
| 120 |
244964_at |
ATCG00580
|
PSBE, photosystem II reaction center protein E |
-0.586 |
-0.010 |
| 121 |
256354_at |
AT1G54870
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.585 |
0.030 |
| 122 |
244979_at |
ATCG00750
|
RPS11, ribosomal protein S11 |
-0.579 |
0.000 |
| 123 |
249271_at |
AT5G41790
|
CIP1, COP1-interactive protein 1 |
-0.576 |
-0.092 |
| 124 |
245005_at |
ATCG00330
|
RPS14, chloroplast ribosomal protein S14 |
-0.573 |
0.028 |
| 125 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-0.563 |
0.038 |
| 126 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
-0.562 |
0.183 |
| 127 |
245019_at |
ATCG00530
|
YCF10 |
-0.560 |
-0.005 |
| 128 |
244984_at |
ATCG00800
|
[RESISTANCE TO PSEUDOMONAS SYRINGAE 3 (RPS3)] |
-0.559 |
-0.005 |
| 129 |
260166_at |
AT1G79840
|
GL2, GLABRA 2 |
-0.553 |
0.092 |
| 130 |
245010_at |
ATCG00420
|
NDHJ, NADH dehydrogenase subunit J |
-0.551 |
-0.058 |
| 131 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
-0.550 |
0.006 |
| 132 |
249045_at |
AT5G44380
|
[FAD-binding Berberine family protein] |
-0.549 |
0.024 |
| 133 |
254828_at |
AT4G12550
|
AIR1, Auxin-Induced in Root cultures 1 |
-0.534 |
-0.013 |
| 134 |
250500_at |
AT5G09530
|
PELPK1, Pro-Glu-Leu|Ile|Val-Pro-Lys 1, PRP10, proline-rich protein 10 |
-0.529 |
0.102 |
| 135 |
245004_at |
ATCG00300
|
YCF9 |
-0.528 |
-0.034 |
| 136 |
251942_at |
AT3G53480
|
ABCG37, ATP-binding cassette G37, ATPDR9, PLEIOTROPIC DRUG RESISTANCE 9, PDR9, pleiotropic drug resistance 9, PIS1, polar auxin transport inhibitor sensitive 1 |
-0.522 |
-0.027 |
| 137 |
245009_at |
ATCG00380
|
RPS4, chloroplast ribosomal protein S4 |
-0.521 |
-0.024 |
| 138 |
244997_at |
ATCG00170
|
RPOC2 |
-0.520 |
-0.047 |
| 139 |
244943_at |
ATMG00070
|
NAD9, NADH dehydrogenase subunit 9 |
-0.518 |
-0.024 |
| 140 |
253191_at |
AT4G35350
|
XCP1, xylem cysteine peptidase 1 |
-0.518 |
0.066 |
| 141 |
247297_at |
AT5G64100
|
[Peroxidase superfamily protein] |
-0.515 |
-0.005 |
| 142 |
245018_at |
ATCG00520
|
YCF4 |
-0.510 |
-0.013 |
| 143 |
244986_at |
ATCG00820
|
RPS19, ribosomal protein S19 |
-0.502 |
-0.009 |
| 144 |
245015_at |
ATCG00490
|
RBCL |
-0.497 |
-0.085 |
| 145 |
247437_at |
AT5G62490
|
ATHVA22B, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE B, HVA22B, HVA22 homologue B |
-0.493 |
-0.043 |
| 146 |
248728_at |
AT5G48000
|
CYP708 A2, CYTOCHROME P450, FAMILY 708, SUBFAMILY A, POLYPEPTIDE 2, CYP708A2, cytochrome P450, family 708, subfamily A, polypeptide 2, THAH, THALIANOL HYDROXYLASE, THAH1, THALIANOL HYDROXYLASE 1 |
-0.493 |
-0.031 |
| 147 |
249542_at |
AT5G38140
|
NF-YC12, nuclear factor Y, subunit C12 |
-0.492 |
0.005 |
| 148 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
-0.485 |
-0.050 |
| 149 |
250868_at |
AT5G03860
|
MLS, malate synthase |
-0.484 |
-0.044 |
| 150 |
267459_at |
AT2G33850
|
unknown |
-0.483 |
0.616 |
| 151 |
245024_at |
ATCG00120
|
ATPA, ATP synthase subunit alpha |
-0.482 |
-0.024 |
| 152 |
257333_at |
ATMG01360
|
COX1, cytochrome oxidase |
-0.481 |
-0.003 |
| 153 |
260088_at |
AT1G73190
|
ALPHA-TIP, ALPHA-TONOPLAST INTRINSIC PROTEIN, TIP3;1 |
-0.475 |
-0.072 |
| 154 |
262260_at |
AT1G70850
|
MLP34, MLP-like protein 34 |
-0.475 |
0.102 |
| 155 |
245000_at |
ATCG00210
|
YCF6 |
-0.469 |
-0.022 |
| 156 |
259578_at |
AT1G27990
|
unknown |
-0.468 |
-0.045 |
| 157 |
244969_at |
ATCG00650
|
RPS18, ribosomal protein S18 |
-0.461 |
-0.007 |
| 158 |
245006_at |
ATCG00340
|
PSAB |
-0.458 |
-0.018 |
| 159 |
249567_at |
AT5G38020
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.458 |
-0.026 |