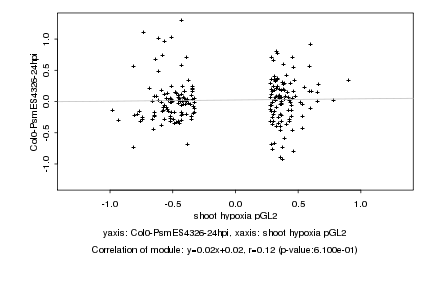
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
shoot hypoxia pGL2 |
Log2 signal ratio
Col0-PsmES4326-24hpi |
| 1 |
245385_at |
AT4G14020
|
[Rapid alkalinization factor (RALF) family protein] |
0.895 |
0.342 |
| 2 |
260672_at |
AT1G19480
|
[DNA glycosylase superfamily protein] |
0.775 |
0.032 |
| 3 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
0.658 |
0.279 |
| 4 |
250881_at |
AT5G04080
|
unknown |
0.650 |
0.151 |
| 5 |
250646_at |
AT5G06720
|
ATPA2, peroxidase 2, AtPRX53, PA2, peroxidase 2, PRX53, peroxidase 53 |
0.648 |
0.004 |
| 6 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
0.604 |
0.173 |
| 7 |
264580_at |
AT1G05340
|
unknown |
0.597 |
0.922 |
| 8 |
259332_at |
AT3G03830
|
SAUR28, SMALL AUXIN UP RNA 28 |
0.597 |
-0.108 |
| 9 |
259423_at |
AT1G13880
|
[ELM2 domain-containing protein] |
0.587 |
0.176 |
| 10 |
265025_at |
AT1G24575
|
unknown |
0.583 |
0.564 |
| 11 |
246653_at |
AT5G35200
|
[ENTH/ANTH/VHS superfamily protein] |
0.549 |
0.225 |
| 12 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
0.532 |
-0.042 |
| 13 |
255028_at |
AT4G09890
|
unknown |
0.530 |
-0.421 |
| 14 |
249583_at |
AT5G37770
|
CML24, CALMODULIN-LIKE 24, TCH2, TOUCH 2 |
0.528 |
-0.231 |
| 15 |
250650_at |
AT5G06850
|
FTIP1, FT-interacting protein 1 |
0.518 |
-0.003 |
| 16 |
259329_at |
AT3G16360
|
AHP4, HPT phosphotransmitter 4 |
0.472 |
0.083 |
| 17 |
246440_at |
AT5G17650
|
[glycine/proline-rich protein] |
0.466 |
0.342 |
| 18 |
261576_at |
AT1G01070
|
UMAMIT28, Usually multiple acids move in and out Transporters 28 |
0.462 |
0.173 |
| 19 |
260887_at |
AT1G29160
|
[Dof-type zinc finger DNA-binding family protein] |
0.459 |
0.559 |
| 20 |
247252_at |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
0.457 |
-0.797 |
| 21 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
0.451 |
-0.449 |
| 22 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
0.451 |
0.706 |
| 23 |
255945_at |
AT5G28610
|
unknown |
0.449 |
0.053 |
| 24 |
246108_at |
AT5G28630
|
[glycine-rich protein] |
0.443 |
-0.141 |
| 25 |
246518_at |
AT5G15770
|
AtGNA1, glucose-6-phosphate acetyltransferase 1, GNA1, glucose-6-phosphate acetyltransferase 1 |
0.442 |
-0.226 |
| 26 |
266479_at |
AT2G31160
|
LSH3, LIGHT SENSITIVE HYPOCOTYLS 3, OBO1, ORGAN BOUNDARY 1 |
0.442 |
-0.053 |
| 27 |
255243_at |
AT4G05590
|
unknown |
0.434 |
0.298 |
| 28 |
255854_at |
AT1G67050
|
unknown |
0.432 |
-0.003 |
| 29 |
261407_at |
AT1G18810
|
[phytochrome kinase substrate-related] |
0.428 |
-0.287 |
| 30 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.428 |
-0.308 |
| 31 |
251454_at |
AT3G60080
|
[RING/U-box superfamily protein] |
0.424 |
0.030 |
| 32 |
248466_at |
AT5G50720
|
ATHVA22E, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE E, HVA22E, HVA22 homologue E |
0.421 |
-0.137 |
| 33 |
249622_at |
AT5G37550
|
unknown |
0.418 |
0.070 |
| 34 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
0.415 |
0.152 |
| 35 |
263297_at |
AT2G15310
|
ARFB1A, ADP-ribosylation factor B1A, ATARFB1A, ADP-ribosylation factor B1A |
0.406 |
0.420 |
| 36 |
258529_at |
AT3G06740
|
GATA15, GATA transcription factor 15 |
0.404 |
-0.353 |
| 37 |
254361_at |
AT4G22212
|
[Arabidopsis defensin-like protein] |
0.392 |
0.096 |
| 38 |
262357_at |
AT1G73040
|
[Mannose-binding lectin superfamily protein] |
0.391 |
0.292 |
| 39 |
252765_at |
AT3G42800
|
unknown |
0.390 |
0.046 |
| 40 |
254685_at |
AT4G13790
|
SAUR25, SMALL AUXIN UPREGULATED RNA 25 |
0.388 |
0.190 |
| 41 |
260805_at |
AT1G78320
|
ATGSTU23, glutathione S-transferase TAU 23, GSTU23, glutathione S-transferase TAU 23 |
0.386 |
-0.579 |
| 42 |
258091_at |
AT3G14560
|
unknown |
0.381 |
0.284 |
| 43 |
251790_at |
AT3G55470
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.380 |
0.602 |
| 44 |
253243_at |
AT4G34560
|
unknown |
0.377 |
0.201 |
| 45 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
0.374 |
-0.914 |
| 46 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
0.373 |
-0.735 |
| 47 |
261409_at |
AT1G07640
|
OBP2, URP3, UAS-TAGGED ROOT PATTERNING3 |
0.370 |
0.310 |
| 48 |
263782_at |
AT2G46380
|
unknown |
0.366 |
-0.219 |
| 49 |
251197_at |
AT3G62960
|
[Thioredoxin superfamily protein] |
0.365 |
-0.032 |
| 50 |
257698_at |
AT3G12730
|
[Homeodomain-like superfamily protein] |
0.362 |
0.003 |
| 51 |
266322_at |
AT2G46690
|
SAUR32, SMALL AUXIN UPREGULATED RNA 32 |
0.362 |
0.077 |
| 52 |
256527_at |
AT1G66100
|
[Plant thionin] |
0.362 |
-0.027 |
| 53 |
250741_at |
AT5G05790
|
[Duplicated homeodomain-like superfamily protein] |
0.361 |
-0.313 |
| 54 |
265357_at |
AT2G16740
|
UBC29, ubiquitin-conjugating enzyme 29 |
0.360 |
0.178 |
| 55 |
248028_at |
AT5G55620
|
unknown |
0.359 |
-0.893 |
| 56 |
254362_at |
AT4G22160
|
unknown |
0.358 |
-0.044 |
| 57 |
257365_x_at |
AT2G26020
|
PDF1.2b, plant defensin 1.2b |
0.355 |
-0.459 |
| 58 |
266175_at |
AT2G02450
|
ANAC034, Arabidopsis NAC domain containing protein 34, ANAC035, NAC domain containing protein 35, AtLOV1, LOV1, LONG VEGETATIVE PHASE 1, NAC035, NAC domain containing protein 35 |
0.355 |
-0.201 |
| 59 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
0.354 |
-0.387 |
| 60 |
265769_at |
AT2G48090
|
unknown |
0.351 |
0.208 |
| 61 |
260004_at |
AT1G67860
|
unknown |
0.350 |
0.105 |
| 62 |
257245_at |
AT3G24110
|
[Calcium-binding EF-hand family protein] |
0.350 |
0.060 |
| 63 |
247632_at |
AT5G60460
|
[Preprotein translocase Sec, Sec61-beta subunit protein] |
0.347 |
0.094 |
| 64 |
251154_at |
AT3G63110
|
ATIPT3, isopentenyltransferase 3, IPT3, isopentenyltransferase 3 |
0.341 |
-0.350 |
| 65 |
248967_at |
AT5G45350
|
[proline-rich family protein] |
0.338 |
0.090 |
| 66 |
256880_at |
AT3G26450
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.338 |
-0.250 |
| 67 |
257058_at |
AT3G15352
|
ATCOX17, ARABIDOPSIS THALIANA CYTOCHROME C OXIDASE 17, COX17, cytochrome c oxidase 17 |
0.333 |
0.375 |
| 68 |
249532_at |
AT5G38780
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.331 |
0.781 |
| 69 |
267364_at |
AT2G40080
|
ELF4, EARLY FLOWERING 4 |
0.331 |
0.355 |
| 70 |
256596_at |
AT3G28540
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.328 |
0.252 |
| 71 |
264782_at |
AT1G08810
|
AtMYB60, myb domain protein 60, MYB60, myb domain protein 60 |
0.325 |
-0.384 |
| 72 |
252740_at |
AT3G43270
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.324 |
0.217 |
| 73 |
252870_at |
AT4G39940
|
AKN2, APS-kinase 2, APK2, ADENOSINE-5'-PHOSPHOSULFATE (APS) KINASE 2 |
0.323 |
-0.084 |
| 74 |
249166_at |
AT5G42840
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.321 |
0.093 |
| 75 |
257517_at |
AT3G16330
|
unknown |
0.320 |
0.808 |
| 76 |
249811_at |
AT5G23760
|
[Copper transport protein family] |
0.315 |
0.023 |
| 77 |
254559_at |
AT4G19200
|
[proline-rich family protein] |
0.313 |
0.065 |
| 78 |
262314_at |
AT1G70810
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.313 |
0.352 |
| 79 |
265354_at |
AT2G16700
|
ADF5, actin depolymerizing factor 5, ATADF5 |
0.312 |
0.336 |
| 80 |
250223_at |
AT5G14070
|
ROXY2 |
0.312 |
0.030 |
| 81 |
253162_at |
AT4G35630
|
PSAT1, phosphoserine aminotransferase 1 |
0.311 |
0.409 |
| 82 |
259765_at |
AT1G64370
|
unknown |
0.310 |
0.035 |
| 83 |
265926_at |
AT2G18600
|
[Ubiquitin-conjugating enzyme family protein] |
0.310 |
0.355 |
| 84 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
0.309 |
-0.660 |
| 85 |
253726_at |
AT4G29430
|
rps15ae, ribosomal protein S15A E |
0.309 |
-0.154 |
| 86 |
264899_at |
AT1G23130
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.308 |
0.034 |
| 87 |
265825_at |
AT2G35635
|
RUB2, RELATED TO UBIQUITIN 2, UBQ7, ubiquitin 7 |
0.304 |
0.180 |
| 88 |
252712_at |
AT3G43800
|
ATGSTU27, glutathione S-transferase tau 27, GSTU27, glutathione S-transferase tau 27 |
0.300 |
-0.196 |
| 89 |
261500_at |
AT1G28400
|
unknown |
0.298 |
-0.314 |
| 90 |
248230_at |
AT5G53830
|
[VQ motif-containing protein] |
0.297 |
0.669 |
| 91 |
259250_at |
AT3G07580
|
unknown |
0.297 |
0.256 |
| 92 |
262880_at |
AT1G64880
|
[Ribosomal protein S5 family protein] |
0.295 |
-0.365 |
| 93 |
263656_at |
AT1G04240
|
IAA3, indole-3-acetic acid inducible 3, SHY2, SHORT HYPOCOTYL 2 |
0.293 |
-0.768 |
| 94 |
259426_at |
AT1G01470
|
LEA14, LATE EMBRYOGENESIS ABUNDANT 14, LSR3, LIGHT STRESS-REGULATED 3 |
0.292 |
0.162 |
| 95 |
259366_at |
AT1G13280
|
AOC4, allene oxide cyclase 4 |
0.291 |
-0.250 |
| 96 |
246585_at |
AT5G14750
|
ATMYB66, myb domain protein 66, MYB66, myb domain protein 66, WER, WEREWOLF, WER1, WEREWOLF 1 |
0.290 |
0.000 |
| 97 |
256131_at |
AT1G13600
|
AtbZIP58, basic leucine-zipper 58, bZIP58, basic leucine-zipper 58 |
0.290 |
-0.012 |
| 98 |
264893_at |
AT1G23140
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.287 |
0.357 |
| 99 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
0.286 |
0.720 |
| 100 |
247656_at |
AT5G59890
|
ADF4, actin depolymerizing factor 4, ATADF4 |
0.286 |
0.194 |
| 101 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.283 |
-0.151 |
| 102 |
258006_at |
AT3G19400
|
[Cysteine proteinases superfamily protein] |
0.282 |
0.127 |
| 103 |
257635_at |
AT3G26280
|
CYP71B4, cytochrome P450, family 71, subfamily B, polypeptide 4 |
0.280 |
-0.051 |
| 104 |
261375_at |
AT1G53160
|
FTM6, FLORAL TRANSITION AT THE MERISTEM6, SPL4, squamosa promoter binding protein-like 4 |
0.280 |
-0.686 |
| 105 |
249878_at |
AT5G23090
|
NF-YB13, nuclear factor Y, subunit B13 |
0.278 |
0.303 |
| 106 |
264318_at |
AT1G04220
|
KCS2, 3-ketoacyl-CoA synthase 2 |
0.277 |
-0.061 |
| 107 |
250471_at |
AT5G10170
|
ATMIPS3, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 3, MIPS3, myo-inositol-1-phosphate synthase 3 |
0.277 |
-0.309 |
| 108 |
263047_at |
AT2G17630
|
PSAT2, phosphoserine aminotransferase 2 |
0.276 |
-0.112 |
| 109 |
260532_at |
AT2G47330
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.274 |
0.080 |
| 110 |
244981_at |
ATCG00770
|
RPS8, ribosomal protein S8 |
-0.988 |
-0.131 |
| 111 |
244966_at |
ATCG00600
|
PETG |
-0.938 |
-0.295 |
| 112 |
245049_at |
ATCG00050
|
RPS16, ribosomal protein S16 |
-0.820 |
-0.731 |
| 113 |
266544_at |
AT2G35300
|
AtLEA4-2, late embryogenesis abundant 4-2, LEA18, late embryogenesis abundant 18, LEA4-2, late embryogenesis abundant 4-2 |
-0.813 |
0.567 |
| 114 |
244985_at |
ATCG00810
|
RPL22, ribosomal protein L22 |
-0.807 |
-0.208 |
| 115 |
245025_at |
ATCG00130
|
ATPF |
-0.781 |
-0.200 |
| 116 |
244980_at |
ATCG00760
|
RPL36, ribosomal protein L36 |
-0.767 |
-0.144 |
| 117 |
245008_at |
ATCG00360
|
YCF3 |
-0.765 |
-0.311 |
| 118 |
245016_at |
ATCG00500
|
ACCD, acetyl-CoA carboxylase carboxyl transferase subunit beta |
-0.745 |
-0.283 |
| 119 |
244970_at |
ATCG00660
|
RPL20, ribosomal protein L20 |
-0.742 |
-0.242 |
| 120 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
-0.734 |
1.114 |
| 121 |
252019_at |
AT3G53040
|
[late embryogenesis abundant protein, putative / LEA protein, putative] |
-0.693 |
0.213 |
| 122 |
244964_at |
ATCG00580
|
PSBE, photosystem II reaction center protein E |
-0.664 |
0.002 |
| 123 |
244932_at |
ATCG01060
|
PSAC |
-0.664 |
-0.275 |
| 124 |
245011_at |
ATCG00430
|
PSBG, photosystem II reaction center protein G |
-0.657 |
-0.440 |
| 125 |
245005_at |
ATCG00330
|
RPS14, chloroplast ribosomal protein S14 |
-0.653 |
0.086 |
| 126 |
244983_at |
ATCG00790
|
RPL16, ribosomal protein L16 |
-0.651 |
-0.231 |
| 127 |
245002_at |
ATCG00270
|
PSBD, photosystem II reaction center protein D |
-0.647 |
-0.212 |
| 128 |
244982_at |
ATCG00780
|
RPL14, ribosomal protein L14 |
-0.646 |
-0.173 |
| 129 |
249045_at |
AT5G44380
|
[FAD-binding Berberine family protein] |
-0.640 |
0.686 |
| 130 |
257130_at |
AT3G20210
|
DELTA-VPE, delta vacuolar processing enzyme, DELTAVPE, DELTA VACUOLAR PROCESSING ENZYME |
-0.634 |
0.090 |
| 131 |
244965_at |
ATCG00590
|
ORF31 |
-0.620 |
0.020 |
| 132 |
258347_at |
AT3G17520
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.618 |
0.481 |
| 133 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.614 |
1.024 |
| 134 |
250868_at |
AT5G03860
|
MLS, malate synthase |
-0.597 |
-0.004 |
| 135 |
244995_at |
ATCG00150
|
ATPI |
-0.595 |
-0.373 |
| 136 |
247297_at |
AT5G64100
|
[Peroxidase superfamily protein] |
-0.591 |
0.179 |
| 137 |
244933_at |
ATCG01070
|
NDHE |
-0.584 |
-0.149 |
| 138 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
-0.582 |
0.742 |
| 139 |
244936_at |
ATCG01100
|
NDHA |
-0.580 |
-0.087 |
| 140 |
245017_at |
ATCG00510
|
PSAI, photsystem I subunit I |
-0.576 |
-0.138 |
| 141 |
247437_at |
AT5G62490
|
ATHVA22B, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE B, HVA22B, HVA22 homologue B |
-0.569 |
0.125 |
| 142 |
249101_at |
AT5G43580
|
UPI, UNUSUAL SERINE PROTEASE INHIBITOR |
-0.568 |
0.969 |
| 143 |
266654_at |
AT2G25890
|
[Oleosin family protein] |
-0.567 |
-0.052 |
| 144 |
244934_at |
ATCG01080
|
NDHG |
-0.567 |
-0.281 |
| 145 |
244975_at |
ATCG00710
|
PSBH, photosystem II reaction center protein H |
-0.555 |
-0.087 |
| 146 |
256354_at |
AT1G54870
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.554 |
0.060 |
| 147 |
249353_at |
AT5G40420
|
OLE2, OLEOSIN 2, OLEO2, oleosin 2 |
-0.549 |
0.138 |
| 148 |
244979_at |
ATCG00750
|
RPS11, ribosomal protein S11 |
-0.547 |
-0.122 |
| 149 |
244963_at |
ATCG00570
|
PSBF, photosystem II reaction center protein F |
-0.537 |
0.032 |
| 150 |
245018_at |
ATCG00520
|
YCF4 |
-0.537 |
-0.159 |
| 151 |
254828_at |
AT4G12550
|
AIR1, Auxin-Induced in Root cultures 1 |
-0.536 |
0.029 |
| 152 |
249271_at |
AT5G41790
|
CIP1, COP1-interactive protein 1 |
-0.524 |
-0.014 |
| 153 |
244967_at |
ATCG00630
|
PSAJ |
-0.523 |
0.052 |
| 154 |
244937_at |
ATCG01110
|
NDHH, NAD(P)H dehydrogenase subunit H |
-0.522 |
-0.265 |
| 155 |
244998_at |
ATCG00180
|
RPOC1 |
-0.522 |
-0.239 |
| 156 |
244997_at |
ATCG00170
|
RPOC2 |
-0.518 |
-0.319 |
| 157 |
245021_at |
ATCG00550
|
PSBJ, photosystem II reaction center protein J |
-0.517 |
0.035 |
| 158 |
245019_at |
ATCG00530
|
YCF10 |
-0.515 |
-0.015 |
| 159 |
258224_at |
AT3G15670
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.513 |
0.244 |
| 160 |
246273_at |
AT4G36700
|
[RmlC-like cupins superfamily protein] |
-0.513 |
1.032 |
| 161 |
245003_at |
ATCG00280
|
PSBC, photosystem II reaction center protein C |
-0.511 |
-0.162 |
| 162 |
245022_at |
ATCG00560
|
PSBL, photosystem II reaction center protein L |
-0.505 |
0.013 |
| 163 |
264698_at |
AT1G70200
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.495 |
-0.278 |
| 164 |
244996_at |
ATCG00160
|
RPS2, ribosomal protein S2 |
-0.491 |
-0.341 |
| 165 |
244984_at |
ATCG00800
|
[RESISTANCE TO PSEUDOMONAS SYRINGAE 3 (RPS3)] |
-0.489 |
-0.172 |
| 166 |
248352_at |
AT5G52300
|
LTI65, LOW-TEMPERATURE-INDUCED 65, RD29B, RESPONSIVE TO DESSICATION 29B |
-0.485 |
0.156 |
| 167 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
-0.480 |
0.153 |
| 168 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
-0.473 |
0.132 |
| 169 |
245010_at |
ATCG00420
|
NDHJ, NADH dehydrogenase subunit J |
-0.471 |
-0.325 |
| 170 |
245015_at |
ATCG00490
|
RBCL |
-0.460 |
-0.318 |
| 171 |
253035_at |
AT4G38240
|
CGL, COMPLEX GLYCAN LESS, CGL1, COMPLEX GLYCAN LESS 1, GNTI, N-ACETYLGLUCOSAMINYLTRANSFERASE I |
-0.459 |
0.111 |
| 172 |
245000_at |
ATCG00210
|
YCF6 |
-0.459 |
-0.022 |
| 173 |
256814_at |
AT3G21370
|
BGLU19, beta glucosidase 19 |
-0.459 |
0.072 |
| 174 |
257333_at |
ATMG01360
|
COX1, cytochrome oxidase |
-0.458 |
0.028 |
| 175 |
245026_at |
ATCG00140
|
ATPH |
-0.451 |
-0.336 |
| 176 |
251838_at |
AT3G54940
|
[Papain family cysteine protease] |
-0.448 |
0.051 |
| 177 |
260088_at |
AT1G73190
|
ALPHA-TIP, ALPHA-TONOPLAST INTRINSIC PROTEIN, TIP3;1 |
-0.445 |
0.014 |
| 178 |
266143_at |
AT2G38905
|
[Low temperature and salt responsive protein family] |
-0.443 |
0.004 |
| 179 |
245009_at |
ATCG00380
|
RPS4, chloroplast ribosomal protein S4 |
-0.437 |
-0.168 |
| 180 |
249304_at |
AT5G41480
|
ATDFA, A. THALIANA DHFS-FPGS HOMOLOG A, DFA, DHFS-FPGS HOMOLOG A, EMB9, EMBRYO DEFECTIVE 9, GLA1, GLOBULAR ARREST1 |
-0.437 |
-0.048 |
| 181 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
-0.437 |
0.577 |
| 182 |
260716_at |
AT1G48130
|
ATPER1, 1-cysteine peroxiredoxin 1, PER1, 1-cysteine peroxiredoxin 1 |
-0.437 |
0.114 |
| 183 |
264166_at |
AT1G65370
|
[TRAF-like family protein] |
-0.434 |
-0.294 |
| 184 |
258957_at |
AT3G01420
|
ALPHA-DOX1, alpha-dioxygenase 1, DIOX1, DOX1, PADOX-1, plant alpha dioxygenase 1 |
-0.432 |
1.313 |
| 185 |
245024_at |
ATCG00120
|
ATPA, ATP synthase subunit alpha |
-0.431 |
-0.213 |
| 186 |
245004_at |
ATCG00300
|
YCF9 |
-0.430 |
0.012 |
| 187 |
254247_at |
AT4G23260
|
CRK18, cysteine-rich RLK (RECEPTOR-like protein kinase) 18 |
-0.430 |
0.241 |
| 188 |
244986_at |
ATCG00820
|
RPS19, ribosomal protein S19 |
-0.424 |
-0.195 |
| 189 |
245014_at |
ATCG00480
|
ATPB, ATP synthase subunit beta, PB, ATP synthase subunit beta |
-0.416 |
-0.046 |
| 190 |
247672_at |
AT5G60220
|
TET4, tetraspanin4 |
-0.415 |
0.067 |
| 191 |
256815_at |
AT3G21380
|
[Mannose-binding lectin superfamily protein] |
-0.412 |
0.054 |
| 192 |
244943_at |
ATMG00070
|
NAD9, NADH dehydrogenase subunit 9 |
-0.409 |
0.167 |
| 193 |
251942_at |
AT3G53480
|
ABCG37, ATP-binding cassette G37, ATPDR9, PLEIOTROPIC DRUG RESISTANCE 9, PDR9, pleiotropic drug resistance 9, PIS1, polar auxin transport inhibitor sensitive 1 |
-0.409 |
0.169 |
| 194 |
244960_at |
ATCG01020
|
RPL32, ribosomal protein L32 |
-0.405 |
0.026 |
| 195 |
258863_at |
AT3G03300
|
ATDCL2, DICER-LIKE 2, DCL2, dicer-like 2 |
-0.400 |
-0.045 |
| 196 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
-0.396 |
0.710 |
| 197 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
-0.394 |
0.015 |
| 198 |
249144_at |
AT5G43270
|
SPL2, squamosa promoter binding protein-like 2 |
-0.391 |
-0.192 |
| 199 |
261782_at |
AT1G76110
|
[HMG (high mobility group) box protein with ARID/BRIGHT DNA-binding domain] |
-0.384 |
-0.685 |
| 200 |
257322_at |
ATMG01180
|
ORF111B |
-0.384 |
0.037 |
| 201 |
251137_at |
AT5G01300
|
[PEBP (phosphatidylethanolamine-binding protein) family protein] |
-0.382 |
0.343 |
| 202 |
244935_at |
ATCG01090
|
NDHI |
-0.375 |
-0.019 |
| 203 |
244976_at |
ATCG00720
|
PETB, photosynthetic electron transfer B |
-0.360 |
-0.037 |
| 204 |
249542_at |
AT5G38140
|
NF-YC12, nuclear factor Y, subunit C12 |
-0.353 |
-0.238 |
| 205 |
245555_at |
AT4G15390
|
[HXXXD-type acyl-transferase family protein] |
-0.351 |
0.107 |
| 206 |
244968_at |
ATCG00640
|
RPL33, ribosomal protein L33 |
-0.351 |
-0.272 |
| 207 |
244906_at |
ATMG00690
|
ORF240A |
-0.350 |
0.037 |
| 208 |
266690_at |
AT2G19900
|
ATNADP-ME1, Arabidopsis thaliana NADP-malic enzyme 1, NADP-ME1, NADP-malic enzyme 1 |
-0.348 |
0.212 |
| 209 |
264835_at |
AT1G03550
|
AtSCAMP4, SCAMP4, Secretory carrier membrane protein 4 |
-0.347 |
0.244 |
| 210 |
252208_at |
AT3G50380
|
[INVOLVED IN: protein localization] |
-0.346 |
0.162 |
| 211 |
256099_at |
AT1G13710
|
CYP78A5, cytochrome P450, family 78, subfamily A, polypeptide 5, KLU, KLUH |
-0.343 |
0.205 |
| 212 |
244961_at |
ATCG01040
|
YCF5 |
-0.342 |
-0.072 |
| 213 |
244962_at |
ATCG01050
|
NDHD |
-0.342 |
-0.176 |
| 214 |
264469_at |
AT1G67100
|
LBD40, LOB domain-containing protein 40 |
-0.334 |
0.060 |
| 215 |
245047_at |
ATCG00020
|
PSBA, photosystem II reaction center protein A |
-0.333 |
-0.015 |
| 216 |
255310_at |
AT4G04955
|
ALN, allantoinase, ATALN, allantoinase |
-0.332 |
-0.173 |
| 217 |
263726_at |
AT2G13610
|
ABCG5, ATP-binding cassette G5 |
-0.332 |
-0.098 |