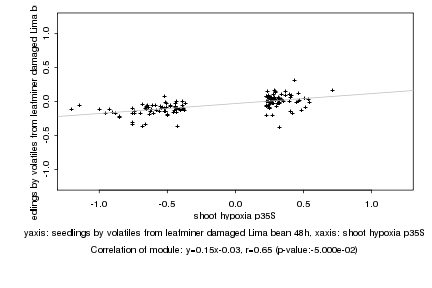
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
shoot hypoxia p35S |
Log2 signal ratio
seedlings by volatiles from leafminer damaged Lima bean 48h |
| 1 |
245385_at |
AT4G14020
|
[Rapid alkalinization factor (RALF) family protein] |
0.712 |
0.177 |
| 2 |
257714_at |
AT3G27360
|
H3.1, histone 3.1 |
0.537 |
-0.004 |
| 3 |
249253_at |
AT5G42060
|
[DEK, chromatin associated protein] |
0.530 |
0.038 |
| 4 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
0.510 |
-0.077 |
| 5 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
0.501 |
0.051 |
| 6 |
264580_at |
AT1G05340
|
unknown |
0.482 |
-0.119 |
| 7 |
265909_at |
AT2G25720
|
unknown |
0.465 |
0.025 |
| 8 |
246861_at |
AT5G25890
|
IAA28, indole-3-acetic acid inducible 28, IAR2, IAA-ALANINE RESISTANT 2 |
0.459 |
0.125 |
| 9 |
246518_at |
AT5G15770
|
AtGNA1, glucose-6-phosphate acetyltransferase 1, GNA1, glucose-6-phosphate acetyltransferase 1 |
0.456 |
0.012 |
| 10 |
249583_at |
AT5G37770
|
CML24, CALMODULIN-LIKE 24, TCH2, TOUCH 2 |
0.444 |
-0.003 |
| 11 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.431 |
0.323 |
| 12 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
0.413 |
-0.167 |
| 13 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
0.410 |
0.101 |
| 14 |
247252_at |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
0.404 |
-0.135 |
| 15 |
260887_at |
AT1G29160
|
[Dof-type zinc finger DNA-binding family protein] |
0.402 |
0.061 |
| 16 |
264059_at |
AT2G31305
|
INH3, INHIBITOR-3 |
0.395 |
0.011 |
| 17 |
254909_at |
AT4G11210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.392 |
0.107 |
| 18 |
257365_x_at |
AT2G26020
|
PDF1.2b, plant defensin 1.2b |
0.365 |
0.103 |
| 19 |
257058_at |
AT3G15352
|
ATCOX17, ARABIDOPSIS THALIANA CYTOCHROME C OXIDASE 17, COX17, cytochrome c oxidase 17 |
0.361 |
0.149 |
| 20 |
247747_at |
AT5G59000
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.346 |
0.009 |
| 21 |
253726_at |
AT4G29430
|
rps15ae, ribosomal protein S15A E |
0.342 |
0.016 |
| 22 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
0.334 |
0.115 |
| 23 |
253830_at |
AT4G27652
|
unknown |
0.332 |
0.036 |
| 24 |
256527_at |
AT1G66100
|
[Plant thionin] |
0.323 |
-0.374 |
| 25 |
257245_at |
AT3G24110
|
[Calcium-binding EF-hand family protein] |
0.322 |
-0.006 |
| 26 |
266322_at |
AT2G46690
|
SAUR32, SMALL AUXIN UPREGULATED RNA 32 |
0.322 |
-0.024 |
| 27 |
251985_at |
AT3G53220
|
[Thioredoxin superfamily protein] |
0.318 |
0.048 |
| 28 |
265025_at |
AT1G24575
|
unknown |
0.314 |
0.065 |
| 29 |
258091_at |
AT3G14560
|
unknown |
0.313 |
0.052 |
| 30 |
249878_at |
AT5G23090
|
NF-YB13, nuclear factor Y, subunit B13 |
0.309 |
-0.017 |
| 31 |
260672_at |
AT1G19480
|
[DNA glycosylase superfamily protein] |
0.309 |
0.029 |
| 32 |
256880_at |
AT3G26450
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.304 |
-0.026 |
| 33 |
252457_at |
AT3G47180
|
[RING/U-box superfamily protein] |
0.297 |
-0.063 |
| 34 |
245367_at |
AT4G16265
|
NRPB9B, NRPD9B, NRPE9B |
0.294 |
0.020 |
| 35 |
253607_at |
AT4G30330
|
[Small nuclear ribonucleoprotein family protein] |
0.291 |
0.031 |
| 36 |
267364_at |
AT2G40080
|
ELF4, EARLY FLOWERING 4 |
0.289 |
0.142 |
| 37 |
258002_at |
AT3G28930
|
AIG2, AVRRPT2-INDUCED GENE 2 |
0.287 |
0.153 |
| 38 |
251790_at |
AT3G55470
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.286 |
0.064 |
| 39 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.281 |
0.172 |
| 40 |
264895_at |
AT1G23100
|
[GroES-like family protein] |
0.278 |
0.104 |
| 41 |
266566_at |
AT2G24040
|
[Low temperature and salt responsive protein family] |
0.274 |
-0.020 |
| 42 |
249441_at |
AT5G39730
|
[AIG2-like (avirulence induced gene) family protein] |
0.273 |
0.059 |
| 43 |
261407_at |
AT1G18810
|
[phytochrome kinase substrate-related] |
0.272 |
-0.206 |
| 44 |
248028_at |
AT5G55620
|
unknown |
0.271 |
0.016 |
| 45 |
251393_at |
AT3G60640
|
ATG8G, AUTOPHAGY 8G |
0.267 |
-0.015 |
| 46 |
247111_at |
AT5G65880
|
unknown |
0.261 |
0.061 |
| 47 |
261409_at |
AT1G07640
|
OBP2, URP3, UAS-TAGGED ROOT PATTERNING3 |
0.256 |
-0.000 |
| 48 |
257929_at |
AT3G16980
|
NRPB9A, NRPD9A, NRPE9A |
0.255 |
-0.003 |
| 49 |
259250_at |
AT3G07580
|
unknown |
0.255 |
0.014 |
| 50 |
265354_at |
AT2G16700
|
ADF5, actin depolymerizing factor 5, ATADF5 |
0.253 |
0.066 |
| 51 |
259661_at |
AT1G55265
|
unknown |
0.252 |
0.033 |
| 52 |
254756_at |
AT4G13235
|
EDA21, embryo sac development arrest 21 |
0.247 |
0.048 |
| 53 |
247292_at |
AT5G64490
|
[ARM repeat superfamily protein] |
0.246 |
-0.078 |
| 54 |
254355_at |
AT4G22380
|
[Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein] |
0.246 |
0.068 |
| 55 |
254624_at |
AT4G18580
|
unknown |
0.245 |
0.065 |
| 56 |
266717_at |
AT2G46735
|
unknown |
0.244 |
-0.093 |
| 57 |
267636_at |
AT2G42110
|
unknown |
0.243 |
-0.094 |
| 58 |
249834_at |
AT5G23440
|
FTRA1, ferredoxin/thioredoxin reductase subunit A (variable subunit) 1 |
0.243 |
-0.040 |
| 59 |
265676_at |
AT2G32070
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
0.239 |
0.011 |
| 60 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
0.237 |
-0.051 |
| 61 |
249427_at |
AT5G39850
|
[Ribosomal protein S4] |
0.236 |
0.095 |
| 62 |
265357_at |
AT2G16740
|
UBC29, ubiquitin-conjugating enzyme 29 |
0.235 |
0.061 |
| 63 |
262314_at |
AT1G70810
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.233 |
0.157 |
| 64 |
252661_at |
AT3G44450
|
unknown |
0.230 |
0.002 |
| 65 |
262236_at |
AT1G48330
|
unknown |
0.226 |
-0.052 |
| 66 |
250434_at |
AT5G10390
|
H3.1, histone 3.1 |
0.226 |
0.001 |
| 67 |
265494_at |
AT2G15680
|
AtCML30, CML30, calmodulin-like 30 |
0.225 |
-0.072 |
| 68 |
255243_at |
AT4G05590
|
unknown |
0.225 |
0.081 |
| 69 |
255511_at |
AT4G02075
|
PIT1, pitchoun 1 |
0.224 |
-0.206 |
| 70 |
255011_at |
AT4G10040
|
CYTC-2, cytochrome c-2 |
0.223 |
0.087 |
| 71 |
244981_at |
ATCG00770
|
RPS8, ribosomal protein S8 |
-1.209 |
-0.109 |
| 72 |
244985_at |
ATCG00810
|
RPL22, ribosomal protein L22 |
-1.146 |
-0.057 |
| 73 |
244980_at |
ATCG00760
|
RPL36, ribosomal protein L36 |
-1.004 |
-0.107 |
| 74 |
245025_at |
ATCG00130
|
ATPF |
-0.956 |
-0.174 |
| 75 |
245049_at |
ATCG00050
|
RPS16, ribosomal protein S16 |
-0.931 |
-0.106 |
| 76 |
244970_at |
ATCG00660
|
RPL20, ribosomal protein L20 |
-0.904 |
-0.158 |
| 77 |
245008_at |
ATCG00360
|
YCF3 |
-0.887 |
-0.176 |
| 78 |
245016_at |
ATCG00500
|
ACCD, acetyl-CoA carboxylase carboxyl transferase subunit beta |
-0.858 |
-0.207 |
| 79 |
244966_at |
ATCG00600
|
PETG |
-0.853 |
-0.222 |
| 80 |
244983_at |
ATCG00790
|
RPL16, ribosomal protein L16 |
-0.763 |
-0.091 |
| 81 |
244933_at |
ATCG01070
|
NDHE |
-0.763 |
-0.329 |
| 82 |
244932_at |
ATCG01060
|
PSAC |
-0.759 |
-0.300 |
| 83 |
244982_at |
ATCG00780
|
RPL14, ribosomal protein L14 |
-0.758 |
-0.162 |
| 84 |
245017_at |
ATCG00510
|
PSAI, photsystem I subunit I |
-0.745 |
-0.145 |
| 85 |
245002_at |
ATCG00270
|
PSBD, photosystem II reaction center protein D |
-0.743 |
-0.176 |
| 86 |
245011_at |
ATCG00430
|
PSBG, photosystem II reaction center protein G |
-0.698 |
-0.170 |
| 87 |
244934_at |
ATCG01080
|
NDHG |
-0.689 |
-0.362 |
| 88 |
244984_at |
ATCG00800
|
[RESISTANCE TO PSEUDOMONAS SYRINGAE 3 (RPS3)] |
-0.684 |
-0.040 |
| 89 |
245003_at |
ATCG00280
|
PSBC, photosystem II reaction center protein C |
-0.665 |
-0.099 |
| 90 |
245005_at |
ATCG00330
|
RPS14, chloroplast ribosomal protein S14 |
-0.665 |
-0.334 |
| 91 |
244998_at |
ATCG00180
|
RPOC1 |
-0.659 |
-0.078 |
| 92 |
244996_at |
ATCG00160
|
RPS2, ribosomal protein S2 |
-0.659 |
-0.109 |
| 93 |
244975_at |
ATCG00710
|
PSBH, photosystem II reaction center protein H |
-0.655 |
-0.059 |
| 94 |
244964_at |
ATCG00580
|
PSBE, photosystem II reaction center protein E |
-0.649 |
-0.058 |
| 95 |
245019_at |
ATCG00530
|
YCF10 |
-0.645 |
-0.079 |
| 96 |
244995_at |
ATCG00150
|
ATPI |
-0.631 |
-0.178 |
| 97 |
244936_at |
ATCG01100
|
NDHA |
-0.627 |
-0.144 |
| 98 |
244997_at |
ATCG00170
|
RPOC2 |
-0.621 |
-0.106 |
| 99 |
244979_at |
ATCG00750
|
RPS11, ribosomal protein S11 |
-0.611 |
-0.048 |
| 100 |
244965_at |
ATCG00590
|
ORF31 |
-0.608 |
-0.169 |
| 101 |
244963_at |
ATCG00570
|
PSBF, photosystem II reaction center protein F |
-0.588 |
-0.053 |
| 102 |
245018_at |
ATCG00520
|
YCF4 |
-0.583 |
-0.130 |
| 103 |
245026_at |
ATCG00140
|
ATPH |
-0.562 |
-0.137 |
| 104 |
245004_at |
ATCG00300
|
YCF9 |
-0.552 |
-0.065 |
| 105 |
244967_at |
ATCG00630
|
PSAJ |
-0.543 |
-0.092 |
| 106 |
245000_at |
ATCG00210
|
YCF6 |
-0.538 |
-0.080 |
| 107 |
245015_at |
ATCG00490
|
RBCL |
-0.527 |
-0.094 |
| 108 |
254247_at |
AT4G23260
|
CRK18, cysteine-rich RLK (RECEPTOR-like protein kinase) 18 |
-0.522 |
0.081 |
| 109 |
245024_at |
ATCG00120
|
ATPA, ATP synthase subunit alpha |
-0.521 |
-0.147 |
| 110 |
257333_at |
ATMG01360
|
COX1, cytochrome oxidase |
-0.519 |
-0.011 |
| 111 |
245010_at |
ATCG00420
|
NDHJ, NADH dehydrogenase subunit J |
-0.519 |
-0.146 |
| 112 |
244986_at |
ATCG00820
|
RPS19, ribosomal protein S19 |
-0.516 |
-0.138 |
| 113 |
249271_at |
AT5G41790
|
CIP1, COP1-interactive protein 1 |
-0.513 |
-0.025 |
| 114 |
245009_at |
ATCG00380
|
RPS4, chloroplast ribosomal protein S4 |
-0.511 |
-0.074 |
| 115 |
244937_at |
ATCG01110
|
NDHH, NAD(P)H dehydrogenase subunit H |
-0.506 |
-0.179 |
| 116 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-0.505 |
-0.193 |
| 117 |
244943_at |
ATMG00070
|
NAD9, NADH dehydrogenase subunit 9 |
-0.480 |
-0.046 |
| 118 |
264835_at |
AT1G03550
|
AtSCAMP4, SCAMP4, Secretory carrier membrane protein 4 |
-0.479 |
-0.060 |
| 119 |
250572_at |
AT5G08210
|
MIR834A, microRNA834A |
-0.461 |
-0.148 |
| 120 |
251641_at |
AT3G57470
|
[Insulinase (Peptidase family M16) family protein] |
-0.451 |
-0.131 |
| 121 |
245006_at |
ATCG00340
|
PSAB |
-0.446 |
-0.064 |
| 122 |
258863_at |
AT3G03300
|
ATDCL2, DICER-LIKE 2, DCL2, dicer-like 2 |
-0.446 |
-0.059 |
| 123 |
245014_at |
ATCG00480
|
ATPB, ATP synthase subunit beta, PB, ATP synthase subunit beta |
-0.445 |
-0.024 |
| 124 |
244976_at |
ATCG00720
|
PETB, photosynthetic electron transfer B |
-0.443 |
-0.088 |
| 125 |
244903_at |
ATMG00660
|
ORF149 |
-0.439 |
0.004 |
| 126 |
259573_at |
AT1G20390
|
[gypsy-like retrotransposon family, has a 1.5e-251 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
-0.435 |
-0.109 |
| 127 |
249542_at |
AT5G38140
|
NF-YC12, nuclear factor Y, subunit C12 |
-0.433 |
-0.157 |
| 128 |
244962_at |
ATCG01050
|
NDHD |
-0.430 |
-0.361 |
| 129 |
244935_at |
ATCG01090
|
NDHI |
-0.430 |
-0.092 |
| 130 |
255044_at |
AT4G09680
|
ATCTC1, CTC1, conserved telomere maintenance component 1 |
-0.414 |
-0.104 |
| 131 |
244968_at |
ATCG00640
|
RPL33, ribosomal protein L33 |
-0.410 |
-0.124 |
| 132 |
244906_at |
ATMG00690
|
ORF240A |
-0.393 |
0.005 |
| 133 |
245020_at |
ATCG00540
|
PETA, photosynthetic electron transfer A |
-0.389 |
-0.050 |
| 134 |
245047_at |
ATCG00020
|
PSBA, photosystem II reaction center protein A |
-0.389 |
-0.103 |
| 135 |
265902_at |
AT2G25590
|
[Plant Tudor-like protein] |
-0.389 |
-0.086 |
| 136 |
244961_at |
ATCG01040
|
YCF5 |
-0.385 |
-0.100 |
| 137 |
249006_at |
AT5G44660
|
unknown |
-0.383 |
-0.114 |
| 138 |
245926_at |
AT5G24740
|
SHBY, SHRUBBY |
-0.381 |
-0.131 |
| 139 |
247564_at |
AT5G61140
|
[U5 small nuclear ribonucleoprotein helicase] |
-0.372 |
-0.018 |