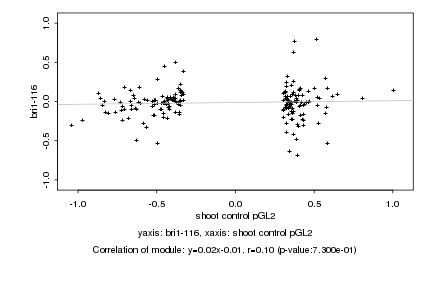
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
shoot control pGL2 |
Log2 signal ratio
bri1-116 |
| 1 |
245385_at |
AT4G14020
|
[Rapid alkalinization factor (RALF) family protein] |
1.003 |
0.146 |
| 2 |
260672_at |
AT1G19480
|
[DNA glycosylase superfamily protein] |
0.803 |
0.046 |
| 3 |
250650_at |
AT5G06850
|
FTIP1, FT-interacting protein 1 |
0.642 |
0.102 |
| 4 |
259423_at |
AT1G13880
|
[ELM2 domain-containing protein] |
0.613 |
0.073 |
| 5 |
250646_at |
AT5G06720
|
ATPA2, peroxidase 2, AtPRX53, PA2, peroxidase 2, PRX53, peroxidase 53 |
0.581 |
-0.529 |
| 6 |
260805_at |
AT1G78320
|
ATGSTU23, glutathione S-transferase TAU 23, GSTU23, glutathione S-transferase TAU 23 |
0.580 |
0.172 |
| 7 |
259549_at |
AT1G35290
|
ALT1, acyl-lipid thioesterase 1 |
0.573 |
-0.066 |
| 8 |
260076_at |
AT1G73630
|
[EF hand calcium-binding protein family] |
0.567 |
0.301 |
| 9 |
246653_at |
AT5G35200
|
[ENTH/ANTH/VHS superfamily protein] |
0.566 |
-0.143 |
| 10 |
246429_at |
AT5G17450
|
HIPP21, heavy metal associated isoprenylated plant protein 21 |
0.529 |
0.042 |
| 11 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
0.524 |
-0.273 |
| 12 |
249583_at |
AT5G37770
|
CML24, CALMODULIN-LIKE 24, TCH2, TOUCH 2 |
0.521 |
-0.045 |
| 13 |
252650_at |
AT3G44690
|
unknown |
0.517 |
0.064 |
| 14 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
0.514 |
0.799 |
| 15 |
265025_at |
AT1G24575
|
unknown |
0.502 |
0.178 |
| 16 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
0.470 |
0.006 |
| 17 |
249126_at |
AT5G43380
|
TOPP6, type one serine/threonine protein phosphatase 6 |
0.463 |
0.131 |
| 18 |
263297_at |
AT2G15310
|
ARFB1A, ADP-ribosylation factor B1A, ATARFB1A, ADP-ribosylation factor B1A |
0.458 |
-0.009 |
| 19 |
246440_at |
AT5G17650
|
[glycine/proline-rich protein] |
0.447 |
-0.009 |
| 20 |
258880_at |
AT3G06420
|
ATG8H, autophagy 8h |
0.433 |
-0.037 |
| 21 |
252133_at |
AT3G50900
|
unknown |
0.430 |
-0.155 |
| 22 |
255243_at |
AT4G05590
|
unknown |
0.430 |
-0.299 |
| 23 |
262196_at |
AT1G77870
|
MUB5, membrane-anchored ubiquitin-fold protein 5 precursor |
0.429 |
-0.044 |
| 24 |
255028_at |
AT4G09890
|
unknown |
0.427 |
-0.241 |
| 25 |
246270_at |
AT4G36500
|
unknown |
0.423 |
-0.225 |
| 26 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
0.420 |
0.079 |
| 27 |
245990_at |
AT5G20640
|
unknown |
0.418 |
0.000 |
| 28 |
261407_at |
AT1G18810
|
[phytochrome kinase substrate-related] |
0.413 |
0.167 |
| 29 |
248466_at |
AT5G50720
|
ATHVA22E, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE E, HVA22E, HVA22 homologue E |
0.409 |
-0.171 |
| 30 |
258091_at |
AT3G14560
|
unknown |
0.407 |
-0.038 |
| 31 |
261881_at |
AT1G80760
|
NIP6, NIP6;1, NOD26-like intrinsic protein 6;1, NLM7 |
0.404 |
0.160 |
| 32 |
251454_at |
AT3G60080
|
[RING/U-box superfamily protein] |
0.403 |
0.148 |
| 33 |
252866_at |
AT4G39840
|
unknown |
0.402 |
-0.060 |
| 34 |
257698_at |
AT3G12730
|
[Homeodomain-like superfamily protein] |
0.400 |
0.085 |
| 35 |
258617_at |
AT3G03000
|
[EF hand calcium-binding protein family] |
0.395 |
-0.314 |
| 36 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
0.391 |
-0.680 |
| 37 |
251790_at |
AT3G55470
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.390 |
-0.286 |
| 38 |
261576_at |
AT1G01070
|
UMAMIT28, Usually multiple acids move in and out Transporters 28 |
0.389 |
0.023 |
| 39 |
247252_at |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
0.385 |
-0.479 |
| 40 |
262880_at |
AT1G64880
|
[Ribosomal protein S5 family protein] |
0.384 |
-0.005 |
| 41 |
261286_at |
AT1G35780
|
unknown |
0.382 |
0.016 |
| 42 |
262357_at |
AT1G73040
|
[Mannose-binding lectin superfamily protein] |
0.382 |
0.041 |
| 43 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
0.377 |
0.085 |
| 44 |
263890_at |
AT2G37030
|
SAUR46, SMALL AUXIN UPREGULATED RNA 46 |
0.374 |
0.773 |
| 45 |
259616_at |
AT1G47960
|
ATC/VIF1, CELL WALL / VACUOLAR INHIBITOR OF FRUCTOSIDASE 1, C/VIF1, cell wall / vacuolar inhibitor of fructosidase 1 |
0.371 |
0.006 |
| 46 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
0.368 |
-0.412 |
| 47 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
0.368 |
0.265 |
| 48 |
248230_at |
AT5G53830
|
[VQ motif-containing protein] |
0.367 |
-0.117 |
| 49 |
246518_at |
AT5G15770
|
AtGNA1, glucose-6-phosphate acetyltransferase 1, GNA1, glucose-6-phosphate acetyltransferase 1 |
0.366 |
0.124 |
| 50 |
262092_at |
AT1G56150
|
SAUR71, SMALL AUXIN UPREGULATED 71 |
0.365 |
0.631 |
| 51 |
263067_at |
AT2G17550
|
TRM26, TON1 Recruiting Motif 26 |
0.362 |
-0.223 |
| 52 |
246585_at |
AT5G14750
|
ATMYB66, myb domain protein 66, MYB66, myb domain protein 66, WER, WEREWOLF, WER1, WEREWOLF 1 |
0.362 |
-0.145 |
| 53 |
265926_at |
AT2G18600
|
[Ubiquitin-conjugating enzyme family protein] |
0.360 |
-0.081 |
| 54 |
265769_at |
AT2G48090
|
unknown |
0.358 |
0.095 |
| 55 |
263047_at |
AT2G17630
|
PSAT2, phosphoserine aminotransferase 2 |
0.354 |
-0.028 |
| 56 |
254201_at |
AT4G24130
|
unknown |
0.351 |
-0.122 |
| 57 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
0.351 |
-0.222 |
| 58 |
251197_at |
AT3G62960
|
[Thioredoxin superfamily protein] |
0.350 |
-0.008 |
| 59 |
254362_at |
AT4G22160
|
unknown |
0.350 |
0.210 |
| 60 |
260662_at |
AT1G19540
|
[NmrA-like negative transcriptional regulator family protein] |
0.348 |
0.070 |
| 61 |
252958_at |
AT4G38620
|
ATMYB4, myb domain protein 4, MYB4, myb domain protein 4 |
0.348 |
-0.028 |
| 62 |
259756_at |
AT1G71080
|
[RNA polymerase II transcription elongation factor] |
0.348 |
-0.030 |
| 63 |
266326_at |
AT2G46650
|
ATCB5-C, ARABIDOPSIS CYTOCHROME B5 ISOFORM C, B5 #1, CB5-C, cytochrome B5 isoform C, CYTB5-C |
0.344 |
-0.014 |
| 64 |
252765_at |
AT3G42800
|
unknown |
0.342 |
-0.625 |
| 65 |
247656_at |
AT5G59890
|
ADF4, actin depolymerizing factor 4, ATADF4 |
0.341 |
-0.005 |
| 66 |
250515_at |
AT5G09570
|
[Cox19-like CHCH family protein] |
0.340 |
0.005 |
| 67 |
259832_at |
AT1G69580
|
[Homeodomain-like superfamily protein] |
0.339 |
-0.070 |
| 68 |
258879_at |
AT3G03270
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.339 |
-0.053 |
| 69 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
0.333 |
-0.161 |
| 70 |
256596_at |
AT3G28540
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.331 |
0.015 |
| 71 |
258527_at |
AT3G06850
|
BCE2, DIN3, DARK INDUCIBLE 3, LTA1 |
0.331 |
-0.082 |
| 72 |
264872_at |
AT1G24260
|
AGL9, AGAMOUS-like 9, SEP3, SEPALLATA3 |
0.328 |
0.070 |
| 73 |
260012_at |
AT1G67865
|
unknown |
0.327 |
0.331 |
| 74 |
266325_at |
AT2G46630
|
unknown |
0.326 |
-0.033 |
| 75 |
256720_at |
AT2G34140
|
[Dof-type zinc finger DNA-binding family protein] |
0.326 |
-0.012 |
| 76 |
255854_at |
AT1G67050
|
unknown |
0.324 |
0.028 |
| 77 |
251895_at |
AT3G54420
|
ATCHITIV, CHITINASE CLASS IV, ATEP3, homolog of carrot EP3-3 chitinase, CHIV, EP3, homolog of carrot EP3-3 chitinase |
0.324 |
-0.031 |
| 78 |
260004_at |
AT1G67860
|
unknown |
0.323 |
0.204 |
| 79 |
261748_at |
AT1G76070
|
unknown |
0.323 |
-0.270 |
| 80 |
263656_at |
AT1G04240
|
IAA3, indole-3-acetic acid inducible 3, SHY2, SHORT HYPOCOTYL 2 |
0.323 |
-0.386 |
| 81 |
266175_at |
AT2G02450
|
ANAC034, Arabidopsis NAC domain containing protein 34, ANAC035, NAC domain containing protein 35, AtLOV1, LOV1, LONG VEGETATIVE PHASE 1, NAC035, NAC domain containing protein 35 |
0.322 |
0.247 |
| 82 |
247632_at |
AT5G60460
|
[Preprotein translocase Sec, Sec61-beta subunit protein] |
0.320 |
0.051 |
| 83 |
247747_at |
AT5G59000
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.319 |
-0.076 |
| 84 |
248967_at |
AT5G45350
|
[proline-rich family protein] |
0.318 |
0.039 |
| 85 |
259340_at |
AT3G03870
|
unknown |
0.318 |
-0.044 |
| 86 |
254998_at |
AT4G09760
|
[Protein kinase superfamily protein] |
0.318 |
-0.087 |
| 87 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
0.316 |
0.075 |
| 88 |
246034_at |
AT5G08350
|
[GRAM domain-containing protein / ABA-responsive protein-related] |
0.315 |
-0.066 |
| 89 |
259329_at |
AT3G16360
|
AHP4, HPT phosphotransmitter 4 |
0.315 |
0.128 |
| 90 |
248596_at |
AT5G49330
|
ATMYB111, ARABIDOPSIS MYB DOMAIN PROTEIN 111, MYB111, myb domain protein 111, PFG3, PRODUCTION OF FLAVONOL GLYCOSIDES 3 |
0.314 |
0.116 |
| 91 |
263545_at |
AT2G21560
|
unknown |
0.307 |
0.125 |
| 92 |
249783_at |
AT5G24270
|
ATSOS3, SALT OVERLY SENSITIVE 3, CBL4, CALCINEURIN B-LIKE PROTEIN 4, SOS3, SALT OVERLY SENSITIVE 3 |
0.305 |
-0.096 |
| 93 |
260783_at |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
0.304 |
0.113 |
| 94 |
260364_at |
AT1G70560
|
CKRC1, cytokinin induced root curling 1, SAV3, SHADE AVOIDANCE 3, TAA1, tryptophan aminotransferase of Arabidopsis 1, WEI8, WEAK ETHYLENE INSENSITIVE 8 |
0.302 |
-0.203 |
| 95 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
0.300 |
-0.111 |
| 96 |
256731_at |
AT3G30340
|
UMAMIT32, Usually multiple acids move in and out Transporters 32 |
0.300 |
0.020 |
| 97 |
266393_at |
AT2G41260
|
ATM17, M17 |
-1.046 |
-0.302 |
| 98 |
266544_at |
AT2G35300
|
AtLEA4-2, late embryogenesis abundant 4-2, LEA18, late embryogenesis abundant 18, LEA4-2, late embryogenesis abundant 4-2 |
-0.975 |
-0.235 |
| 99 |
244981_at |
ATCG00770
|
RPS8, ribosomal protein S8 |
-0.873 |
0.115 |
| 100 |
265211_at |
AT2G36640
|
ATECP63, embryonic cell protein 63, ECP63, embryonic cell protein 63 |
-0.859 |
0.045 |
| 101 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
-0.846 |
-0.050 |
| 102 |
244966_at |
ATCG00600
|
PETG |
-0.836 |
0.010 |
| 103 |
258347_at |
AT3G17520
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.831 |
-0.129 |
| 104 |
244939_at |
ATCG00065
|
RPS12, RIBOSOMAL PROTEIN S12, RPS12A, ribosomal protein S12A |
-0.810 |
-0.147 |
| 105 |
252019_at |
AT3G53040
|
[late embryogenesis abundant protein, putative / LEA protein, putative] |
-0.770 |
0.026 |
| 106 |
249353_at |
AT5G40420
|
OLE2, OLEOSIN 2, OLEO2, oleosin 2 |
-0.762 |
-0.133 |
| 107 |
250868_at |
AT5G03860
|
MLS, malate synthase |
-0.735 |
-0.012 |
| 108 |
264612_at |
AT1G04560
|
[AWPM-19-like family protein] |
-0.729 |
-0.114 |
| 109 |
256464_at |
AT1G32560
|
AtLEA4-1, Late Embryogenesis Abundant 4-1, LEA4-1, Late Embryogenesis Abundant 4-1 |
-0.723 |
-0.235 |
| 110 |
250624_at |
AT5G07330
|
unknown |
-0.720 |
-0.052 |
| 111 |
245049_at |
ATCG00050
|
RPS16, ribosomal protein S16 |
-0.709 |
0.186 |
| 112 |
244985_at |
ATCG00810
|
RPL22, ribosomal protein L22 |
-0.708 |
-0.092 |
| 113 |
251838_at |
AT3G54940
|
[Papain family cysteine protease] |
-0.683 |
-0.210 |
| 114 |
256354_at |
AT1G54870
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.671 |
0.148 |
| 115 |
247437_at |
AT5G62490
|
ATHVA22B, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE B, HVA22B, HVA22 homologue B |
-0.670 |
0.004 |
| 116 |
264079_at |
AT2G28490
|
[RmlC-like cupins superfamily protein] |
-0.665 |
-0.094 |
| 117 |
247297_at |
AT5G64100
|
[Peroxidase superfamily protein] |
-0.661 |
-0.050 |
| 118 |
244980_at |
ATCG00760
|
RPL36, ribosomal protein L36 |
-0.648 |
0.081 |
| 119 |
245008_at |
ATCG00360
|
YCF3 |
-0.647 |
0.042 |
| 120 |
256814_at |
AT3G21370
|
BGLU19, beta glucosidase 19 |
-0.638 |
-0.084 |
| 121 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
-0.632 |
-0.095 |
| 122 |
249045_at |
AT5G44380
|
[FAD-binding Berberine family protein] |
-0.630 |
-0.494 |
| 123 |
258224_at |
AT3G15670
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.617 |
-0.010 |
| 124 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
-0.613 |
0.190 |
| 125 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
-0.606 |
-0.016 |
| 126 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
-0.588 |
-0.278 |
| 127 |
266143_at |
AT2G38905
|
[Low temperature and salt responsive protein family] |
-0.584 |
0.034 |
| 128 |
249082_at |
AT5G44120
|
ATCRA1, CRUCIFERINA, CRA1, CRUCIFERINA, CRU1 |
-0.568 |
-0.319 |
| 129 |
256338_at |
AT1G72100
|
[late embryogenesis abundant domain-containing protein / LEA domain-containing protein] |
-0.560 |
0.015 |
| 130 |
245017_at |
ATCG00510
|
PSAI, photsystem I subunit I |
-0.532 |
0.013 |
| 131 |
244995_at |
ATCG00150
|
ATPI |
-0.527 |
-0.051 |
| 132 |
260088_at |
AT1G73190
|
ALPHA-TIP, ALPHA-TONOPLAST INTRINSIC PROTEIN, TIP3;1 |
-0.526 |
-0.171 |
| 133 |
245555_at |
AT4G15390
|
[HXXXD-type acyl-transferase family protein] |
-0.519 |
-0.171 |
| 134 |
257334_at |
ATMG01370
|
ORF111D |
-0.517 |
0.020 |
| 135 |
245021_at |
ATCG00550
|
PSBJ, photosystem II reaction center protein J |
-0.517 |
0.019 |
| 136 |
244937_at |
ATCG01110
|
NDHH, NAD(P)H dehydrogenase subunit H |
-0.517 |
-0.031 |
| 137 |
244963_at |
ATCG00570
|
PSBF, photosystem II reaction center protein F |
-0.512 |
0.019 |
| 138 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
-0.508 |
0.014 |
| 139 |
248727_at |
AT5G47990
|
CYP705A5, cytochrome P450, family 705, subfamily A, polypeptide 5, THAD, THALIAN-DIOL DESATURASE, THAD1, THALIAN-DIOL DESATURASE |
-0.501 |
-0.531 |
| 140 |
260890_at |
AT1G29090
|
[Cysteine proteinases superfamily protein] |
-0.499 |
0.281 |
| 141 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
-0.497 |
-0.023 |
| 142 |
245070_at |
AT2G23240
|
AtMT4b, Arabidopsis thaliana metallothionein 4b |
-0.480 |
-0.090 |
| 143 |
266941_at |
AT2G18980
|
[Peroxidase superfamily protein] |
-0.475 |
-0.018 |
| 144 |
244983_at |
ATCG00790
|
RPL16, ribosomal protein L16 |
-0.475 |
-0.091 |
| 145 |
250500_at |
AT5G09530
|
PELPK1, Pro-Glu-Leu|Ile|Val-Pro-Lys 1, PRP10, proline-rich protein 10 |
-0.469 |
0.076 |
| 146 |
244984_at |
ATCG00800
|
[RESISTANCE TO PSEUDOMONAS SYRINGAE 3 (RPS3)] |
-0.463 |
-0.147 |
| 147 |
260716_at |
AT1G48130
|
ATPER1, 1-cysteine peroxiredoxin 1, PER1, 1-cysteine peroxiredoxin 1 |
-0.461 |
-0.201 |
| 148 |
244979_at |
ATCG00750
|
RPS11, ribosomal protein S11 |
-0.460 |
-0.013 |
| 149 |
251137_at |
AT5G01300
|
[PEBP (phosphatidylethanolamine-binding protein) family protein] |
-0.457 |
-0.004 |
| 150 |
265672_at |
AT2G31980
|
AtCYS2, PHYTOCYSTATIN 2, CYS2, PHYTOCYSTATIN 2 |
-0.454 |
-0.016 |
| 151 |
260077_at |
AT1G73620
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.453 |
0.454 |
| 152 |
245022_at |
ATCG00560
|
PSBL, photosystem II reaction center protein L |
-0.453 |
0.006 |
| 153 |
244996_at |
ATCG00160
|
RPS2, ribosomal protein S2 |
-0.440 |
-0.033 |
| 154 |
253392_at |
AT4G32650
|
ATKC1, ARABIDOPSIS THALIANA K+ RECTIFYING CHANNEL 1, AtLKT1, A. thaliana low-K+-tolerant 1, KAT3, potassium channel in Arabidopsis thaliana 3, KC1 |
-0.435 |
-0.214 |
| 155 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
-0.434 |
0.046 |
| 156 |
244960_at |
ATCG01020
|
RPL32, ribosomal protein L32 |
-0.434 |
0.058 |
| 157 |
244936_at |
ATCG01100
|
NDHA |
-0.432 |
0.020 |
| 158 |
251531_at |
AT3G58550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.429 |
-0.076 |
| 159 |
245018_at |
ATCG00520
|
YCF4 |
-0.427 |
-0.020 |
| 160 |
250059_at |
AT5G17820
|
[Peroxidase superfamily protein] |
-0.427 |
0.030 |
| 161 |
247362_at |
AT5G63140
|
ATPAP29, purple acid phosphatase 29, PAP29, PAP29, purple acid phosphatase 29 |
-0.422 |
-0.062 |
| 162 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
-0.421 |
-0.094 |
| 163 |
262605_at |
AT1G15170
|
[MATE efflux family protein] |
-0.418 |
0.053 |
| 164 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
-0.409 |
0.036 |
| 165 |
245026_at |
ATCG00140
|
ATPH |
-0.405 |
0.023 |
| 166 |
247061_at |
AT5G66780
|
unknown |
-0.398 |
0.064 |
| 167 |
264469_at |
AT1G67100
|
LBD40, LOB domain-containing protein 40 |
-0.394 |
0.019 |
| 168 |
245009_at |
ATCG00380
|
RPS4, chloroplast ribosomal protein S4 |
-0.392 |
0.047 |
| 169 |
249144_at |
AT5G43270
|
SPL2, squamosa promoter binding protein-like 2 |
-0.387 |
0.098 |
| 170 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-0.386 |
0.007 |
| 171 |
259615_at |
AT1G47980
|
unknown |
-0.384 |
0.043 |
| 172 |
262838_at |
AT1G14960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.383 |
-0.138 |
| 173 |
251065_at |
AT5G01870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.381 |
0.499 |
| 174 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
-0.377 |
-0.012 |
| 175 |
249006_at |
AT5G44660
|
unknown |
-0.367 |
0.174 |
| 176 |
254247_at |
AT4G23260
|
CRK18, cysteine-rich RLK (RECEPTOR-like protein kinase) 18 |
-0.366 |
-0.035 |
| 177 |
254110_at |
AT4G25260
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.360 |
-0.129 |
| 178 |
255048_at |
AT4G09600
|
GASA3, GAST1 protein homolog 3 |
-0.360 |
0.008 |
| 179 |
250228_at |
AT5G13840
|
CCS52B, cell cycle switch protein 52 B, FZR3, FIZZY-related 3 |
-0.359 |
-0.157 |
| 180 |
245024_at |
ATCG00120
|
ATPA, ATP synthase subunit alpha |
-0.353 |
0.025 |
| 181 |
258424_at |
AT3G16750
|
unknown |
-0.352 |
0.146 |
| 182 |
256734_at |
AT3G29390
|
RIK, RS2-interacting KH protein |
-0.352 |
-0.038 |
| 183 |
244943_at |
ATMG00070
|
NAD9, NADH dehydrogenase subunit 9 |
-0.351 |
0.226 |
| 184 |
245014_at |
ATCG00480
|
ATPB, ATP synthase subunit beta, PB, ATP synthase subunit beta |
-0.351 |
0.004 |
| 185 |
258863_at |
AT3G03300
|
ATDCL2, DICER-LIKE 2, DCL2, dicer-like 2 |
-0.350 |
0.116 |
| 186 |
253971_at |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
-0.347 |
0.083 |
| 187 |
256792_at |
AT3G22150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.343 |
0.132 |
| 188 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
-0.340 |
0.124 |
| 189 |
244962_at |
ATCG01050
|
NDHD |
-0.334 |
0.024 |
| 190 |
245448_at |
AT4G16860
|
RPP4, recognition of peronospora parasitica 4 |
-0.332 |
0.094 |
| 191 |
256099_at |
AT1G13710
|
CYP78A5, cytochrome P450, family 78, subfamily A, polypeptide 5, KLU, KLUH |
-0.332 |
0.393 |