|
probeID |
AGICode |
Annotation |
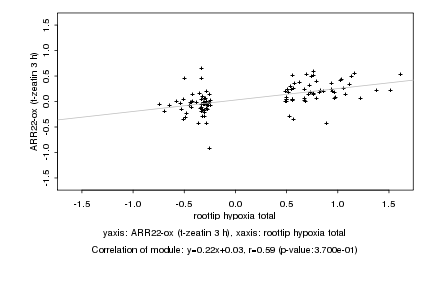
Log2 signal ratio
roottip hypoxia total |
Log2 signal ratio
ARR22-ox (t-zeatin 3 h) |
| 1 |
250464_at |
AT5G10040
|
unknown |
1.609 |
0.546 |
| 2 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
1.514 |
0.231 |
| 3 |
260668_at |
AT1G19530
|
unknown |
1.378 |
0.224 |
| 4 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
1.214 |
0.077 |
| 5 |
254200_at |
AT4G24110
|
unknown |
1.160 |
0.550 |
| 6 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
1.128 |
0.503 |
| 7 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
1.105 |
0.344 |
| 8 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
1.070 |
0.141 |
| 9 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
1.047 |
0.256 |
| 10 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
1.027 |
0.433 |
| 11 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
1.026 |
0.423 |
| 12 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
0.968 |
0.081 |
| 13 |
257153_at |
AT3G27220
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.967 |
0.179 |
| 14 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.960 |
0.072 |
| 15 |
257925_at |
AT3G23170
|
unknown |
0.948 |
0.213 |
| 16 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
0.935 |
0.243 |
| 17 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.932 |
0.357 |
| 18 |
263295_at |
AT2G14210
|
AGL44, AGAMOUS-like 44, ANR1, ARABIDOPSIS NITRATE REGULATED 1 |
0.887 |
-0.423 |
| 19 |
250152_at |
AT5G15120
|
unknown |
0.852 |
0.206 |
| 20 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
0.822 |
0.217 |
| 21 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
0.821 |
0.183 |
| 22 |
261475_at |
AT1G14550
|
[Peroxidase superfamily protein] |
0.784 |
0.397 |
| 23 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
0.784 |
0.075 |
| 24 |
261449_at |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
0.759 |
0.608 |
| 25 |
245076_at |
AT2G23170
|
GH3.3 |
0.757 |
0.513 |
| 26 |
249015_at |
AT5G44730
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.754 |
0.141 |
| 27 |
261567_at |
AT1G33055
|
unknown |
0.754 |
0.165 |
| 28 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.736 |
0.502 |
| 29 |
245998_at |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
0.729 |
0.190 |
| 30 |
260451_at |
AT1G72360
|
AtERF73, ERF73, ethylene response factor 73, HRE1, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 1 |
0.718 |
0.331 |
| 31 |
252882_at |
AT4G39675
|
unknown |
0.712 |
0.142 |
| 32 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.690 |
0.543 |
| 33 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
0.678 |
0.014 |
| 34 |
253722_at |
AT4G29190
|
AtC3H49, AtOZF2, Arabidopsis thaliana oxidation-related zinc finger 2, AtTZF3, OZF2, oxidation-related zinc finger 2, TZF3, tandem zinc finger 3 |
0.670 |
0.076 |
| 35 |
258377_at |
AT3G17690
|
ATCNGC19, CYCLIC NUCLEOTIDE-GATED CHANNEL 19, CNGC19, cyclic nucleotide gated channel 19 |
0.669 |
0.027 |
| 36 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.668 |
0.253 |
| 37 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.622 |
0.379 |
| 38 |
257922_at |
AT3G23150
|
ETR2, ethylene response 2 |
0.569 |
0.372 |
| 39 |
261261_at |
AT1G26730
|
[EXS (ERD1/XPR1/SYG1) family protein] |
0.566 |
0.259 |
| 40 |
248177_at |
AT5G54630
|
[zinc finger protein-related] |
0.563 |
-0.338 |
| 41 |
265501_at |
AT2G15490
|
UGT73B4, UDP-glycosyltransferase 73B4 |
0.556 |
0.523 |
| 42 |
258879_at |
AT3G03270
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.555 |
0.046 |
| 43 |
264561_at |
AT1G55810
|
UKL3, uridine kinase-like 3 |
0.550 |
0.029 |
| 44 |
249234_at |
AT5G42200
|
[RING/U-box superfamily protein] |
0.548 |
0.055 |
| 45 |
263931_at |
AT2G36220
|
unknown |
0.541 |
0.247 |
| 46 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
0.531 |
0.296 |
| 47 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
0.524 |
-0.286 |
| 48 |
259685_at |
AT1G63090
|
AtPP2-A11, phloem protein 2-A11, PP2-A11, phloem protein 2-A11 |
0.515 |
0.186 |
| 49 |
248138_at |
AT5G54960
|
PDC2, pyruvate decarboxylase-2 |
0.508 |
0.237 |
| 50 |
247868_at |
AT5G57620
|
AtMYB36, myb domain protein 36, MYB36, myb domain protein 36 |
0.498 |
0.084 |
| 51 |
252046_at |
AT3G52460
|
[hydroxyproline-rich glycoprotein family protein] |
0.497 |
0.042 |
| 52 |
253404_at |
AT4G32840
|
PFK6, phosphofructokinase 6 |
0.496 |
0.011 |
| 53 |
259681_at |
AT1G77760
|
GNR1, NIA1, nitrate reductase 1, NR1, NITRATE REDUCTASE 1 |
0.491 |
0.057 |
| 54 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
0.486 |
0.197 |
| 55 |
266016_at |
AT2G18670
|
[RING/U-box superfamily protein] |
0.484 |
0.014 |
| 56 |
247914_at |
AT5G57540
|
AtXTH13, XTH13, xyloglucan endotransglucosylase/hydrolase 13 |
-0.751 |
-0.048 |
| 57 |
254056_at |
AT4G25250
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.694 |
-0.186 |
| 58 |
258832_at |
AT3G07070
|
[Protein kinase superfamily protein] |
-0.652 |
-0.078 |
| 59 |
256138_at |
AT1G48670
|
[auxin-responsive GH3 family protein] |
-0.579 |
0.008 |
| 60 |
259195_at |
AT3G01730
|
unknown |
-0.538 |
-0.020 |
| 61 |
253763_at |
AT4G28850
|
ATXTH26, XTH26, xyloglucan endotransglucosylase/hydrolase 26 |
-0.534 |
-0.146 |
| 62 |
261647_at |
AT1G27740
|
RSL4, root hair defective 6-like 4 |
-0.514 |
0.051 |
| 63 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
-0.513 |
-0.340 |
| 64 |
255591_at |
AT4G01630
|
ATEXP17, EXPANSIN 17, ATEXPA17, expansin A17, ATHEXP ALPHA 1.13, EXPA17, expansin A17 |
-0.498 |
0.452 |
| 65 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.496 |
-0.301 |
| 66 |
263376_at |
AT2G20520
|
FLA6, FASCICLIN-like arabinogalactan 6 |
-0.479 |
-0.228 |
| 67 |
263932_at |
AT2G35990
|
LOG2, LONELY GUY 2 |
-0.455 |
-0.064 |
| 68 |
249750_at |
AT5G24570
|
unknown |
-0.447 |
-0.001 |
| 69 |
261241_at |
AT1G32950
|
[Subtilase family protein] |
-0.438 |
-0.003 |
| 70 |
259982_at |
AT1G76410
|
ATL8 |
-0.430 |
-0.117 |
| 71 |
265213_at |
AT1G05020
|
[ENTH/ANTH/VHS superfamily protein] |
-0.425 |
0.149 |
| 72 |
264892_at |
AT1G23160
|
[Auxin-responsive GH3 family protein] |
-0.422 |
0.001 |
| 73 |
260300_at |
AT1G80340
|
ATGA3OX2, ARABIDOPSIS THALIANA GIBBERELLIN-3-OXIDASE 2, GA3OX2, gibberellin 3-oxidase 2, GA4H |
-0.383 |
-0.010 |
| 74 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
-0.368 |
-0.430 |
| 75 |
259525_at |
AT1G12560
|
ATEXP7, ATEXPA7, expansin A7, ATHEXP ALPHA 1.26, EXP7, EXPA7, expansin A7 |
-0.358 |
0.173 |
| 76 |
247871_at |
AT5G57530
|
AtXTH12, XTH12, xyloglucan endotransglucosylase/hydrolase 12 |
-0.342 |
-0.126 |
| 77 |
265411_at |
AT2G16630
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.340 |
-0.043 |
| 78 |
255175_at |
AT4G07960
|
ATCSLC12, Cellulose-synthase-like C12, CSLC12, CELLULOSE-SYNTHASE LIKE C12, CSLC12, Cellulose-synthase-like C12 |
-0.338 |
0.047 |
| 79 |
248183_at |
AT5G54040
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.337 |
0.655 |
| 80 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
-0.337 |
0.466 |
| 81 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.335 |
-0.191 |
| 82 |
265844_at |
AT2G35620
|
FEI2, FEI 2 |
-0.334 |
-0.111 |
| 83 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
-0.328 |
0.108 |
| 84 |
252594_at |
AT3G45680
|
[Major facilitator superfamily protein] |
-0.327 |
-0.279 |
| 85 |
265039_at |
AT1G04000
|
unknown |
-0.326 |
-0.150 |
| 86 |
253723_at |
AT4G29240
|
[Leucine-rich repeat (LRR) family protein] |
-0.323 |
-0.145 |
| 87 |
247498_at |
AT5G61810
|
APC1, ATP/phosphate carrier 1 |
-0.314 |
-0.052 |
| 88 |
267550_at |
AT2G32800
|
AP4.3A, LecRK-S.2, L-type lectin receptor kinase S.2 |
-0.313 |
-0.003 |
| 89 |
254107_at |
AT4G25220
|
AtG3Pp2, glycerol-3-phosphate permease 2, G3Pp2, glycerol-3-phosphate permease 2, RHS15, root hair specific 15 |
-0.312 |
-0.057 |
| 90 |
249934_at |
AT5G22410
|
RHS18, root hair specific 18 |
-0.311 |
-0.293 |
| 91 |
257297_at |
AT3G28040
|
[Leucine-rich receptor-like protein kinase family protein] |
-0.310 |
-0.159 |
| 92 |
265102_at |
AT1G30870
|
[Peroxidase superfamily protein] |
-0.310 |
-0.018 |
| 93 |
260758_at |
AT1G48930
|
AtGH9C1, glycosyl hydrolase 9C1, GH9C1, glycosyl hydrolase 9C1 |
-0.310 |
0.072 |
| 94 |
257131_at |
AT3G20240
|
[Mitochondrial substrate carrier family protein] |
-0.303 |
-0.205 |
| 95 |
262275_at |
AT1G68710
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
-0.295 |
0.061 |
| 96 |
258967_at |
AT3G10470
|
[C2H2-type zinc finger family protein] |
-0.290 |
0.002 |
| 97 |
259846_at |
AT1G72140
|
[Major facilitator superfamily protein] |
-0.288 |
-0.414 |
| 98 |
253100_at |
AT4G37400
|
CYP81F3, cytochrome P450, family 81, subfamily F, polypeptide 3 |
-0.286 |
-0.134 |
| 99 |
251513_at |
AT3G59220
|
ATPIRIN1, PRN, pirin, PRN1 |
-0.285 |
0.214 |
| 100 |
261587_at |
AT1G01660
|
[RING/U-box superfamily protein] |
-0.285 |
-0.030 |
| 101 |
256759_at |
AT3G25640
|
unknown |
-0.284 |
0.004 |
| 102 |
246991_at |
AT5G67400
|
RHS19, root hair specific 19 |
-0.282 |
-0.153 |
| 103 |
255516_at |
AT4G02270
|
RHS13, root hair specific 13 |
-0.279 |
-0.066 |
| 104 |
253734_at |
AT4G29180
|
RHS16, root hair specific 16 |
-0.265 |
-0.075 |
| 105 |
261758_at |
AT1G08250
|
ADT6, arogenate dehydratase 6, AtADT6, Arabidopsis thaliana arogenate dehydratase 6 |
-0.262 |
0.145 |
| 106 |
250744_at |
AT5G05840
|
unknown |
-0.260 |
-0.019 |
| 107 |
263236_at |
AT1G10470
|
ARR4, response regulator 4, ATRR1, RESPONCE REGULATOR 1, IBC7, INDUCED BY CYTOKININ 7, MEE7, maternal effect embryo arrest 7 |
-0.256 |
-0.920 |
| 108 |
266765_at |
AT2G46860
|
AtPPa3, pyrophosphorylase 3, PPa3, pyrophosphorylase 3 |
-0.250 |
-0.077 |
| 109 |
266336_at |
AT2G32270
|
ZIP3, zinc transporter 3 precursor |
-0.249 |
0.023 |