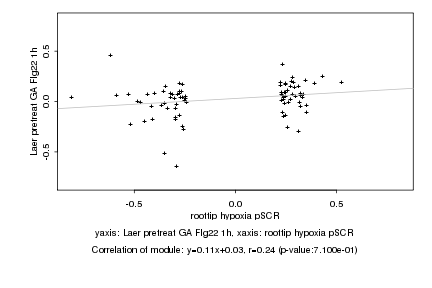
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
roottip hypoxia pSCR |
Log2 signal ratio
Laer pretreat GA Flg22 1h |
| 1 |
246368_at |
AT1G51890
|
[Leucine-rich repeat protein kinase family protein] |
0.520 |
0.196 |
| 2 |
247685_at |
AT5G59680
|
[Leucine-rich repeat protein kinase family protein] |
0.426 |
0.254 |
| 3 |
261052_at |
AT1G01440
|
unknown |
0.389 |
0.184 |
| 4 |
247567_at |
AT5G61190
|
[putative endonuclease or glycosyl hydrolase with C2H2-type zinc finger domain] |
0.349 |
-0.102 |
| 5 |
246765_at |
AT5G27330
|
[Prefoldin chaperone subunit family protein] |
0.347 |
-0.033 |
| 6 |
266627_at |
AT2G35340
|
MEE29, maternal effect embryo arrest 29 |
0.342 |
0.218 |
| 7 |
256176_at |
AT1G51640
|
ATEXO70G2, exocyst subunit exo70 family protein G2, EXO70G2, exocyst subunit exo70 family protein G2 |
0.336 |
0.072 |
| 8 |
262668_at |
AT1G62830
|
ATLSD1, ARABIDOPSIS LYSINE-SPECIFIC HISTONE DEMETHYLASE, ATSWP1, LDL1, LSD1-like 1, LSD1, LYSINE-SPECIFIC HISTONE DEMETHYLASE, SWP1 |
0.329 |
0.043 |
| 9 |
265864_at |
AT2G01750
|
ATMAP70-3, microtubule-associated proteins 70-3, MAP70-3, microtubule-associated proteins 70-3 |
0.318 |
0.087 |
| 10 |
261333_at |
AT1G44910
|
ATPRP40A, pre-mRNA-processing protein 40A, PRP40A, pre-mRNA-processing protein 40A |
0.318 |
0.069 |
| 11 |
259104_at |
AT3G02170
|
LNG2, LONGIFOLIA2, TRM1, TON1 Recruiting Motif 1 |
0.317 |
-0.041 |
| 12 |
251876_at |
AT3G54280
|
ATBTAF1, BTAF1, CHA16, CHR16, RGD3, ROOT GROWTH DEFECTIVE 3 |
0.315 |
-0.007 |
| 13 |
256754_at |
AT3G25690
|
AtCHUP1, Arabidopsis thaliana CHLOROPLAST UNUSUAL POSITIONING 1, CHUP1, CHLOROPLAST UNUSUAL POSITIONING 1 |
0.310 |
-0.293 |
| 14 |
252639_at |
AT3G44550
|
FAR5, fatty acid reductase 5 |
0.308 |
0.159 |
| 15 |
249219_at |
AT5G42400
|
ATXR7, ARABIDOPSIS TRITHORAX-RELATED7, SDG25, SET domain protein 25 |
0.295 |
0.051 |
| 16 |
252047_at |
AT3G52490
|
SMXL3, SMAX1-like 3 |
0.289 |
0.145 |
| 17 |
245166_at |
AT2G33170
|
[Leucine-rich repeat receptor-like protein kinase family protein] |
0.286 |
0.195 |
| 18 |
252395_at |
AT3G47950
|
AHA4, H(+)-ATPase 4, HA4, H(+)-ATPase 4 |
0.282 |
0.075 |
| 19 |
253571_at |
AT4G31000
|
[Calmodulin-binding protein] |
0.282 |
0.239 |
| 20 |
248778_at |
AT5G47940
|
unknown |
0.273 |
0.203 |
| 21 |
246653_at |
AT5G35200
|
[ENTH/ANTH/VHS superfamily protein] |
0.269 |
0.153 |
| 22 |
245588_at |
AT4G15030
|
unknown |
0.268 |
0.025 |
| 23 |
249409_at |
AT5G40340
|
[Tudor/PWWP/MBT superfamily protein] |
0.262 |
-0.008 |
| 24 |
258585_at |
AT3G04340
|
emb2458, embryo defective 2458 |
0.257 |
-0.253 |
| 25 |
262614_at |
AT1G13980
|
EMB30, EMBRYO DEFECTIVE 30, GN, GNOM, VAN7, VASCULAR NETWORK 7 |
0.254 |
0.115 |
| 26 |
249115_at |
AT5G43810
|
AGO10, ARGONAUTE 10, PNH, PINHEAD, ZLL, ZWILLE |
0.247 |
-0.138 |
| 27 |
267015_at |
AT2G39340
|
AtSAC3A, SAC3A, yeast Sac3 homolog A |
0.246 |
0.099 |
| 28 |
264283_at |
AT1G61850
|
[phospholipases] |
0.246 |
0.185 |
| 29 |
258098_at |
AT3G23670
|
KINESIN-12B, PAKRP1L |
0.245 |
0.050 |
| 30 |
256426_at |
AT1G33420
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.244 |
0.173 |
| 31 |
254409_at |
AT4G21400
|
CRK28, cysteine-rich RLK (RECEPTOR-like protein kinase) 28 |
0.239 |
0.090 |
| 32 |
248266_at |
AT5G53440
|
unknown |
0.239 |
-0.015 |
| 33 |
248570_at |
AT5G49780
|
[Leucine-rich repeat protein kinase family protein] |
0.235 |
0.023 |
| 34 |
262956_at |
AT1G54270
|
EIF4A-2, eif4a-2 |
0.234 |
0.044 |
| 35 |
261616_at |
AT1G33060
|
ANAC014, NAC 014, NAC014, NAC 014 |
0.233 |
-0.143 |
| 36 |
251970_at |
AT3G53150
|
UGT73D1, UDP-glucosyl transferase 73D1 |
0.232 |
0.368 |
| 37 |
245575_at |
AT4G14760
|
NET1B, Networked 1B |
0.232 |
-0.102 |
| 38 |
262880_at |
AT1G64880
|
[Ribosomal protein S5 family protein] |
0.228 |
0.062 |
| 39 |
261862_at |
AT1G50410
|
[SNF2 domain-containing protein / helicase domain-containing protein / zinc finger protein-related] |
0.227 |
0.092 |
| 40 |
256864_at |
AT3G23890
|
ATTOPII, TOPII, topoisomerase II |
0.226 |
0.073 |
| 41 |
257285_at |
AT3G29760
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.226 |
0.019 |
| 42 |
258833_at |
AT3G07274
|
|
0.221 |
0.168 |
| 43 |
256282_at |
AT3G12550
|
FDM3, factor of DNA methylation 3 |
0.219 |
0.197 |
| 44 |
247070_at |
AT5G66815
|
unknown |
-0.814 |
0.045 |
| 45 |
254324_at |
AT4G22640
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.620 |
0.460 |
| 46 |
262795_at |
AT1G13130
|
[Cellulase (glycosyl hydrolase family 5) protein] |
-0.590 |
0.064 |
| 47 |
252046_at |
AT3G52460
|
[hydroxyproline-rich glycoprotein family protein] |
-0.532 |
0.071 |
| 48 |
246125_at |
AT5G19875
|
unknown |
-0.520 |
-0.219 |
| 49 |
265463_at |
AT2G37090
|
IRX9, IRREGULAR XYLEM 9 |
-0.485 |
0.003 |
| 50 |
257054_at |
AT3G15353
|
ATMT3, MT3, metallothionein 3 |
-0.473 |
-0.009 |
| 51 |
260389_at |
AT1G74055
|
unknown |
-0.454 |
-0.189 |
| 52 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
-0.436 |
0.078 |
| 53 |
261256_at |
AT1G05760
|
RTM1, restricted tev movement 1 |
-0.417 |
-0.049 |
| 54 |
263689_at |
AT1G26820
|
RNS3, ribonuclease 3 |
-0.414 |
-0.177 |
| 55 |
261019_at |
AT1G26470
|
SNS1, SnRK2-substrate 1 |
-0.403 |
0.088 |
| 56 |
254168_at |
AT4G24250
|
ATMLO13, MILDEW RESISTANCE LOCUS O 13, MLO13, MILDEW RESISTANCE LOCUS O 13 |
-0.369 |
-0.035 |
| 57 |
261317_at |
AT1G53030
|
[Cytochrome C oxidase copper chaperone (COX17)] |
-0.358 |
0.108 |
| 58 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
-0.355 |
-0.507 |
| 59 |
264573_at |
AT1G05310
|
[Pectin lyase-like superfamily protein] |
-0.352 |
-0.014 |
| 60 |
245172_at |
AT2G47540
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.347 |
0.153 |
| 61 |
264288_at |
AT1G62045
|
unknown |
-0.338 |
-0.068 |
| 62 |
249970_at |
AT5G19100
|
[Eukaryotic aspartyl protease family protein] |
-0.325 |
0.044 |
| 63 |
263211_at |
AT1G10460
|
GLP7, germin-like protein 7 |
-0.322 |
0.080 |
| 64 |
255011_at |
AT4G10040
|
CYTC-2, cytochrome c-2 |
-0.316 |
0.074 |
| 65 |
253112_at |
AT4G35970
|
APX5, ascorbate peroxidase 5 |
-0.302 |
0.036 |
| 66 |
257093_at |
AT3G20570
|
AtENODL9, ENODL9, early nodulin-like protein 9 |
-0.300 |
-0.061 |
| 67 |
249214_at |
AT5G42720
|
[Glycosyl hydrolase family 17 protein] |
-0.297 |
-0.152 |
| 68 |
259616_at |
AT1G47960
|
ATC/VIF1, CELL WALL / VACUOLAR INHIBITOR OF FRUCTOSIDASE 1, C/VIF1, cell wall / vacuolar inhibitor of fructosidase 1 |
-0.297 |
-0.178 |
| 69 |
249958_at |
AT5G18970
|
[AWPM-19-like family protein] |
-0.294 |
-0.028 |
| 70 |
247882_at |
AT5G57785
|
unknown |
-0.294 |
-0.637 |
| 71 |
250922_at |
AT5G03345
|
unknown |
-0.288 |
0.073 |
| 72 |
246442_at |
AT5G17610
|
unknown |
-0.281 |
0.081 |
| 73 |
259852_at |
AT1G72280
|
AERO1, endoplasmic reticulum oxidoreductins 1, ERO1, endoplasmic reticulum oxidoreductins 1 |
-0.281 |
0.186 |
| 74 |
265663_at |
AT2G24290
|
unknown |
-0.279 |
-0.134 |
| 75 |
262315_at |
AT1G70990
|
[proline-rich family protein] |
-0.279 |
0.109 |
| 76 |
252296_at |
AT3G48970
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.272 |
0.049 |
| 77 |
246744_at |
AT5G27760
|
[Hypoxia-responsive family protein] |
-0.271 |
0.102 |
| 78 |
263800_at |
AT2G24600
|
[Ankyrin repeat family protein] |
-0.264 |
0.177 |
| 79 |
258866_at |
AT3G03180
|
[Got1/Sft2-like vescicle transport protein family] |
-0.262 |
0.045 |
| 80 |
254187_at |
AT4G23890
|
CRR31, CHLORORESPIRATORY REDUCTION 31, NdhS, NADH dehydrogenase-like complex S |
-0.262 |
-0.240 |
| 81 |
255104_at |
AT4G08685
|
SAH7 |
-0.258 |
-0.271 |
| 82 |
248967_at |
AT5G45350
|
[proline-rich family protein] |
-0.253 |
0.017 |
| 83 |
256424_at |
AT1G33490
|
unknown |
-0.249 |
0.057 |
| 84 |
250079_at |
AT5G16650
|
[Chaperone DnaJ-domain superfamily protein] |
-0.247 |
0.034 |
| 85 |
266783_at |
AT2G29130
|
ATLAC2, LAC2, laccase 2 |
-0.246 |
-0.006 |