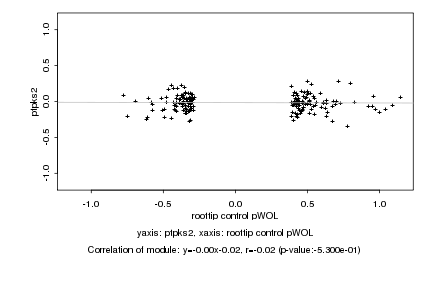
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
roottip control pWOL |
Log2 signal ratio
ptpks2 |
| 1 |
245845_at |
AT1G26150
|
AtPERK10, proline-rich extensin-like receptor kinase 10, PERK10, proline-rich extensin-like receptor kinase 10 |
1.142 |
0.067 |
| 2 |
264145_at |
AT1G79310
|
AtMC7, metacaspase 7, AtMCP2a, metacaspase 2a, MC7, metacaspase 7, MCP2a, metacaspase 2a |
1.089 |
-0.052 |
| 3 |
251044_at |
AT5G02350
|
[Cysteine/Histidine-rich C1 domain family protein] |
1.037 |
-0.100 |
| 4 |
262228_at |
AT1G68690
|
AtPERK9, proline-rich extensin-like receptor kinase 9, PERK9, proline-rich extensin-like receptor kinase 9 |
0.994 |
-0.144 |
| 5 |
253864_at |
AT4G27460
|
CBSX5, CBS domain containing protein 5 |
0.965 |
-0.104 |
| 6 |
257159_at |
AT3G24400
|
AtPERK2, proline-rich extensin-like receptor kinase 2, PERK2, proline-rich extensin-like receptor kinase 2 |
0.956 |
0.070 |
| 7 |
266507_at |
AT2G47860
|
SETH6 |
0.949 |
-0.059 |
| 8 |
250650_at |
AT5G06850
|
FTIP1, FT-interacting protein 1 |
0.920 |
-0.060 |
| 9 |
251229_at |
AT3G62740
|
BGLU7, beta glucosidase 7 |
0.823 |
-0.004 |
| 10 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
0.794 |
0.261 |
| 11 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
0.774 |
-0.337 |
| 12 |
261073_at |
AT1G07300
|
[josephin protein-related] |
0.726 |
-0.022 |
| 13 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
0.710 |
0.284 |
| 14 |
258255_at |
AT3G26800
|
unknown |
0.700 |
0.013 |
| 15 |
260672_at |
AT1G19480
|
[DNA glycosylase superfamily protein] |
0.688 |
-0.041 |
| 16 |
245079_at |
AT2G23330
|
[copia-like retrotransposon family, has a 3.9e-195 P-value blast match to GB:AAA57005 Hopscotch polyprotein (Ty1_Copia-element) (Zea mays)] |
0.680 |
0.003 |
| 17 |
250572_at |
AT5G08210
|
MIR834A, microRNA834A |
0.672 |
-0.061 |
| 18 |
251502_at |
AT3G59130
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.668 |
-0.275 |
| 19 |
264281_at |
AT1G61830
|
[pseudogene] |
0.634 |
-0.151 |
| 20 |
254513_at |
AT4G20235
|
CYP71A28, cytochrome P450, family 71, subfamily A, polypeptide 28 |
0.628 |
0.016 |
| 21 |
245990_at |
AT5G20640
|
unknown |
0.628 |
-0.200 |
| 22 |
253519_at |
AT4G31240
|
AtNRX2, NRX2, nucleoredoxin 2 |
0.627 |
-0.015 |
| 23 |
253432_at |
AT4G32450
|
MEF8S, MEF8 similar |
0.620 |
-0.084 |
| 24 |
252700_at |
AT3G43690
|
[copia-like retrotransposon family protein, has a 1.4e-29 P-value blast match to gb|AAG52950.1| putative envelope protein (Endovir1-1) (Arabidopsis thaliana) (Ty1_Copia-family)] |
0.610 |
-0.003 |
| 25 |
251929_at |
AT3G53920
|
SIG3, SIGMA FACTOR 3, SIGC, RNApolymerase sigma-subunit C |
0.605 |
-0.025 |
| 26 |
260307_at |
AT1G70620
|
[cyclin-related] |
0.590 |
-0.076 |
| 27 |
262880_at |
AT1G64880
|
[Ribosomal protein S5 family protein] |
0.588 |
0.120 |
| 28 |
253506_at |
AT4G31980
|
unknown |
0.557 |
-0.007 |
| 29 |
252569_at |
AT3G45420
|
LecRK-I.4, L-type lectin receptor kinase I.4 |
0.549 |
-0.052 |
| 30 |
267015_at |
AT2G39340
|
AtSAC3A, SAC3A, yeast Sac3 homolog A |
0.543 |
0.050 |
| 31 |
260684_at |
AT1G17590
|
NF-YA8, nuclear factor Y, subunit A8 |
0.542 |
-0.173 |
| 32 |
246653_at |
AT5G35200
|
[ENTH/ANTH/VHS superfamily protein] |
0.540 |
-0.056 |
| 33 |
248452_at |
AT5G51300
|
[splicing factor-related] |
0.536 |
0.071 |
| 34 |
262778_at |
AT1G13050
|
unknown |
0.527 |
-0.110 |
| 35 |
262671_at |
AT1G76040
|
CPK29, calcium-dependent protein kinase 29 |
0.526 |
0.238 |
| 36 |
266529_at |
AT2G16880
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.520 |
-0.029 |
| 37 |
249123_at |
AT5G43760
|
KCS20, 3-ketoacyl-CoA synthase 20 |
0.517 |
0.114 |
| 38 |
248183_at |
AT5G54040
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.513 |
-0.163 |
| 39 |
251076_at |
AT5G01970
|
unknown |
0.510 |
0.016 |
| 40 |
259175_at |
AT3G01560
|
unknown |
0.507 |
0.002 |
| 41 |
267423_at |
AT2G35060
|
KUP11, K+ uptake permease 11 |
0.504 |
0.124 |
| 42 |
248251_at |
AT5G53220
|
unknown |
0.499 |
-0.035 |
| 43 |
255403_at |
AT4G03400
|
DFL2, DWARF IN LIGHT 2, GH3-10 |
0.498 |
0.286 |
| 44 |
259423_at |
AT1G13880
|
[ELM2 domain-containing protein] |
0.495 |
0.145 |
| 45 |
258392_at |
AT3G15400
|
ATA20, anther 20 |
0.493 |
-0.016 |
| 46 |
248060_at |
AT5G55560
|
[Protein kinase superfamily protein] |
0.493 |
0.105 |
| 47 |
248066_at |
AT5G55590
|
QRT1, QUARTET 1 |
0.491 |
0.057 |
| 48 |
265195_at |
AT2G36730
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.485 |
-0.035 |
| 49 |
261382_at |
AT1G05470
|
CVP2, COTYLEDON VASCULAR PATTERN 2 |
0.481 |
0.044 |
| 50 |
264169_at |
AT1G02020
|
[nitroreductase family protein] |
0.478 |
0.129 |
| 51 |
249016_at |
AT5G44750
|
ATREV1, ARABIDOPSIS THALIANA HOMOLOG OF REVERSIONLESS 1, REV1 |
0.471 |
-0.002 |
| 52 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
0.470 |
0.079 |
| 53 |
259666_at |
AT1G55310
|
At-SCL33, SC35-LIKE SPLICING FACTOR 33, ATSCL33, SC35-like splicing factor 33, SCL33, SC35-LIKE SPLICING FACTOR 33, SR33, SC35-like splicing factor 33 |
0.469 |
-0.070 |
| 54 |
248868_at |
AT5G46780
|
[VQ motif-containing protein] |
0.466 |
-0.072 |
| 55 |
263385_at |
AT2G40170
|
ATEM6, ARABIDOPSIS EARLY METHIONINE-LABELLED 6, EM6, EARLY METHIONINE-LABELLED 6, GEA6, LATE EMBRYOGENESIS ABUNDANT 6 |
0.463 |
-0.103 |
| 56 |
249791_at |
AT5G23810
|
AAP7, amino acid permease 7 |
0.463 |
-0.124 |
| 57 |
258196_at |
AT3G13980
|
unknown |
0.462 |
0.006 |
| 58 |
251766_at |
AT3G55910
|
unknown |
0.461 |
-0.024 |
| 59 |
256493_at |
AT1G31600
|
AtTRM9, Arabidopsis thaliana tRNA methyltransferase 9, TRM9, tRNA methyltransferase 9 |
0.453 |
-0.138 |
| 60 |
248369_at |
AT5G52040
|
At-RS41, arginine/serine-rich splicing factor 41, ATRSP41, RS41, arginine/serine-rich splicing factor 41 |
0.452 |
0.144 |
| 61 |
246916_at |
AT5G25910
|
AtRLP52, receptor like protein 52, RLP52, receptor like protein 52 |
0.445 |
-0.166 |
| 62 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
0.444 |
-0.044 |
| 63 |
267627_at |
AT2G42270
|
[U5 small nuclear ribonucleoprotein helicase] |
0.441 |
-0.115 |
| 64 |
253808_at |
AT4G28300
|
unknown |
0.439 |
0.002 |
| 65 |
249974_at |
AT5G18780
|
[F-box/RNI-like superfamily protein] |
0.436 |
-0.020 |
| 66 |
247051_at |
AT5G66670
|
unknown |
0.435 |
-0.097 |
| 67 |
254586_at |
AT4G19510
|
[Disease resistance protein (TIR-NBS-LRR class)] |
0.431 |
0.046 |
| 68 |
255639_at |
AT4G00760
|
APRR8, pseudo-response regulator 8, PRR8, PSEUDO-RESPONSE REGULATOR 8 |
0.430 |
-0.179 |
| 69 |
264257_at |
AT1G09230
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.427 |
0.028 |
| 70 |
253575_at |
AT4G31060
|
[Integrase-type DNA-binding superfamily protein] |
0.427 |
-0.218 |
| 71 |
264437_at |
AT1G27510
|
EX2, EXECUTER 2 |
0.426 |
0.093 |
| 72 |
264513_at |
AT1G09420
|
G6PD4, glucose-6-phosphate dehydrogenase 4 |
0.423 |
-0.204 |
| 73 |
260599_at |
AT1G55940
|
CYP708A1, cytochrome P450, family 708, subfamily A, polypeptide 1 |
0.423 |
-0.019 |
| 74 |
249550_at |
AT5G38210
|
[Protein kinase family protein] |
0.420 |
-0.013 |
| 75 |
262296_at |
AT1G27630
|
CYCT1;3, cyclin T 1;3 |
0.419 |
0.125 |
| 76 |
247813_at |
AT5G58330
|
[lactate/malate dehydrogenase family protein] |
0.419 |
-0.030 |
| 77 |
267075_at |
AT2G41070
|
ATBZIP12, DPBF4, EEL, ENHANCED EM LEVEL |
0.418 |
-0.025 |
| 78 |
262135_at |
AT1G78080
|
AtWIND1, RAP2.4, related to AP2 4, WIND1, wound induced dedifferentiation 1 |
0.417 |
0.042 |
| 79 |
255037_at |
AT4G09460
|
AtMYB6, myb domain protein 6, MYB6, myb domain protein 6 |
0.417 |
-0.057 |
| 80 |
249595_at |
AT5G37930
|
[Protein with RING/U-box and TRAF-like domains] |
0.416 |
0.046 |
| 81 |
251088_at |
AT5G01480
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.415 |
-0.160 |
| 82 |
259242_at |
AT3G33520
|
ARP6, actin-related protein 6, ATARP6, actin-related protein 6, ESD1, EARLY IN SHORT DAYS 1, SUF3, SUPPRESSOR OF FRI 3 |
0.411 |
-0.015 |
| 83 |
251360_at |
AT3G61210
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.407 |
0.004 |
| 84 |
261553_at |
AT1G63420
|
unknown |
0.407 |
0.132 |
| 85 |
265902_at |
AT2G25590
|
[Plant Tudor-like protein] |
0.402 |
-0.048 |
| 86 |
263196_at |
AT1G36070
|
[Transducin/WD40 repeat-like superfamily protein] |
0.401 |
0.092 |
| 87 |
246951_at |
AT5G04880
|
[expressed protein] |
0.401 |
-0.255 |
| 88 |
263260_at |
AT1G10570
|
OTS2, OVERLY TOLERANT TO SALT 2, ULP1C, UB-LIKE PROTEASE 1C |
0.399 |
0.094 |
| 89 |
259000_at |
AT3G01860
|
unknown |
0.397 |
-0.034 |
| 90 |
254188_at |
AT4G23920
|
ATUGE2, UDP-GLC 4-EPIMERASE 2, UGE2, UDP-D-glucose/UDP-D-galactose 4-epimerase 2 |
0.395 |
-0.045 |
| 91 |
252096_at |
AT3G51180
|
[Zinc finger C-x8-C-x5-C-x3-H type family protein] |
0.392 |
-0.144 |
| 92 |
266910_at |
AT2G45920
|
[U-box domain-containing protein] |
0.387 |
-0.198 |
| 93 |
267054_at |
AT2G38370
|
unknown |
0.383 |
-0.011 |
| 94 |
267127_at |
AT2G23610
|
ATMES3, ARABIDOPSIS THALIANA METHYL ESTERASE 3, MES3, methyl esterase 3 |
0.383 |
0.223 |
| 95 |
261232_at |
AT1G20220
|
[Alba DNA/RNA-binding protein] |
-0.783 |
0.095 |
| 96 |
246229_at |
AT4G37160
|
sks15, SKU5 similar 15 |
-0.750 |
-0.208 |
| 97 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
-0.700 |
0.007 |
| 98 |
247094_at |
AT5G66280
|
GMD1, GDP-D-mannose 4,6-dehydratase 1 |
-0.618 |
-0.238 |
| 99 |
254056_at |
AT4G25250
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.616 |
-0.209 |
| 100 |
258432_at |
AT3G16570
|
ATRALF23, ARABIDOPSIS RAPID ALKALINIZATION FACTOR 23, RALF23, rapid alkalinization factor 23 |
-0.606 |
0.047 |
| 101 |
245021_at |
ATCG00550
|
PSBJ, photosystem II reaction center protein J |
-0.586 |
-0.002 |
| 102 |
254606_at |
AT4G19030
|
AT-NLM1, ATNLM1, NOD26-LIKE MAJOR INTRINSIC PROTEIN 1, NIP1;1, NOD26-LIKE INTRINSIC PROTEIN 1;1, NLM1, NOD26-like major intrinsic protein 1 |
-0.579 |
-0.115 |
| 103 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
-0.576 |
-0.036 |
| 104 |
255591_at |
AT4G01630
|
ATEXP17, EXPANSIN 17, ATEXPA17, expansin A17, ATHEXP ALPHA 1.13, EXPA17, expansin A17 |
-0.515 |
0.055 |
| 105 |
260558_at |
AT2G43600
|
[Chitinase family protein] |
-0.506 |
-0.114 |
| 106 |
249756_at |
AT5G24313
|
unknown |
-0.498 |
-0.105 |
| 107 |
259195_at |
AT3G01730
|
unknown |
-0.493 |
-0.214 |
| 108 |
251964_at |
AT3G53370
|
[S1FA-like DNA-binding protein] |
-0.483 |
-0.009 |
| 109 |
262131_at |
AT1G02900
|
ATRALF1, RAPID ALKALINIZATION FACTOR 1, RALF1, rapid alkalinization factor 1, RALFL1, RALF-LIKE 1 |
-0.482 |
0.068 |
| 110 |
255885_at |
AT1G20330
|
CVP1, COTYLEDON VASCULAR PATTERN 1, FRL1, FRILL1, SMT2, sterol methyltransferase 2 |
-0.471 |
0.178 |
| 111 |
246534_at |
AT5G15890
|
TBL21, TRICHOME BIREFRINGENCE-LIKE 21 |
-0.445 |
-0.233 |
| 112 |
265377_at |
AT2G05790
|
[O-Glycosyl hydrolases family 17 protein] |
-0.444 |
0.234 |
| 113 |
263677_at |
AT1G04520
|
PDLP2, plasmodesmata-located protein 2 |
-0.431 |
0.183 |
| 114 |
266687_at |
AT2G19670
|
ATPRMT1A, ARABIDOPSIS THALIANA PROTEIN ARGININE METHYLTRANSFERASE 1A, PRMT1A, protein arginine methyltransferase 1A |
-0.431 |
-0.005 |
| 115 |
247476_at |
AT5G62330
|
unknown |
-0.425 |
-0.044 |
| 116 |
262658_at |
AT1G14220
|
[Ribonuclease T2 family protein] |
-0.423 |
-0.099 |
| 117 |
253763_at |
AT4G28850
|
ATXTH26, XTH26, xyloglucan endotransglucosylase/hydrolase 26 |
-0.421 |
-0.051 |
| 118 |
260549_at |
AT2G43535
|
[Scorpion toxin-like knottin superfamily protein] |
-0.416 |
-0.117 |
| 119 |
250930_at |
AT5G03160
|
ATP58IPK, homolog of mamallian P58IPK, P58IPK, homolog of mamallian P58IPK |
-0.413 |
-0.007 |
| 120 |
265111_at |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.413 |
0.046 |
| 121 |
250108_at |
AT5G15150
|
ATHB-3, homeobox 3, ATHB3, HAT7, HOMEOBOX FROM ARABIDOPSIS THALIANA 7, HB-3, homeobox 3 |
-0.412 |
-0.068 |
| 122 |
250537_at |
AT5G08565
|
[Transcription initiation Spt4-like protein] |
-0.412 |
-0.127 |
| 123 |
261727_at |
AT1G76090
|
SMT3, sterol methyltransferase 3 |
-0.407 |
0.189 |
| 124 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.402 |
0.086 |
| 125 |
266566_at |
AT2G24040
|
[Low temperature and salt responsive protein family] |
-0.393 |
-0.016 |
| 126 |
249469_at |
AT5G39320
|
UDG4, UDP-glucose dehydrogenase 4 |
-0.392 |
0.036 |
| 127 |
264892_at |
AT1G23160
|
[Auxin-responsive GH3 family protein] |
-0.389 |
-0.011 |
| 128 |
262198_at |
AT1G53830
|
ATPME2, pectin methylesterase 2, PME2, pectin methylesterase 2 |
-0.385 |
0.030 |
| 129 |
262978_at |
AT1G75780
|
TUB1, tubulin beta-1 chain |
-0.381 |
0.084 |
| 130 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
-0.376 |
0.236 |
| 131 |
253482_at |
AT4G31985
|
[Ribosomal protein L39 family protein] |
-0.372 |
0.086 |
| 132 |
258345_at |
AT3G22845
|
[emp24/gp25L/p24 family/GOLD family protein] |
-0.370 |
0.057 |
| 133 |
258912_at |
AT3G06460
|
[GNS1/SUR4 membrane protein family] |
-0.370 |
-0.038 |
| 134 |
262864_at |
AT1G64920
|
[UDP-Glycosyltransferase superfamily protein] |
-0.369 |
-0.026 |
| 135 |
250748_at |
AT5G05710
|
[Pleckstrin homology (PH) domain superfamily protein] |
-0.368 |
-0.064 |
| 136 |
261790_at |
AT1G16000
|
unknown |
-0.366 |
0.003 |
| 137 |
253957_at |
AT4G26320
|
AGP13, arabinogalactan protein 13 |
-0.365 |
-0.124 |
| 138 |
249159_at |
AT5G43460
|
[HR-like lesion-inducing protein-related] |
-0.364 |
0.086 |
| 139 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
-0.361 |
-0.018 |
| 140 |
251210_at |
AT3G62810
|
[complex 1 family protein / LVR family protein] |
-0.357 |
0.204 |
| 141 |
265480_at |
AT2G15970
|
ATCOR413-PM1, ARABIDOPSIS THALIANA COLD-REGULATED413 PLASMA MEMBRANE 1, ATCYP19, CYCLOPHILLIN 19, COR413-PM1, cold regulated 413 plasma membrane 1, FL3-5A3, WCOR413, WCOR413-LIKE |
-0.355 |
0.045 |
| 142 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
-0.352 |
0.118 |
| 143 |
247337_at |
AT5G63660
|
LCR74, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 74, PDF2.5 |
-0.351 |
-0.163 |
| 144 |
245172_at |
AT2G47540
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.350 |
-0.090 |
| 145 |
261831_at |
AT1G10630
|
ARFA1F, ADP-ribosylation factor A1F, ATARFA1F, ADP-ribosylation factor A1F |
-0.350 |
-0.053 |
| 146 |
257867_at |
AT3G17780
|
unknown |
-0.347 |
0.130 |
| 147 |
251167_at |
AT3G63360
|
[defensin-related] |
-0.347 |
-0.155 |
| 148 |
251007_at |
AT5G02610
|
[Ribosomal L29 family protein ] |
-0.346 |
0.042 |
| 149 |
247612_at |
AT5G60730
|
[Anion-transporting ATPase] |
-0.346 |
-0.137 |
| 150 |
253914_at |
AT4G27400
|
[Late embryogenesis abundant (LEA) protein-related] |
-0.345 |
-0.154 |
| 151 |
260077_at |
AT1G73620
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.342 |
-0.111 |
| 152 |
254723_at |
AT4G13510
|
AMT1;1, ammonium transporter 1;1, ATAMT1, ARABIDOPSIS THALIANA AMMONIUM TRANSPORT 1, ATAMT1;1 |
-0.341 |
0.051 |
| 153 |
248957_at |
AT5G45620
|
[Proteasome component (PCI) domain protein] |
-0.335 |
0.008 |
| 154 |
250778_at |
AT5G05500
|
MOP10 |
-0.327 |
-0.058 |
| 155 |
256935_at |
AT3G22570
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.327 |
-0.014 |
| 156 |
255279_at |
AT4G04950
|
AtGRXS17, Arabidopsis thaliana monothiol glutaredoxin 17, GRXS17, monothiol glutaredoxin 17 |
-0.326 |
0.112 |
| 157 |
258145_at |
AT3G18200
|
UMAMIT4, Usually multiple acids move in and out Transporters 4 |
-0.326 |
-0.135 |
| 158 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
-0.325 |
-0.124 |
| 159 |
252390_at |
AT3G47836
|
unknown |
-0.324 |
0.007 |
| 160 |
263373_at |
AT2G20515
|
unknown |
-0.324 |
0.013 |
| 161 |
255554_at |
AT4G01897
|
unknown |
-0.324 |
0.060 |
| 162 |
249934_at |
AT5G22410
|
RHS18, root hair specific 18 |
-0.323 |
-0.136 |
| 163 |
259592_at |
AT1G27950
|
LTPG1, glycosylphosphatidylinositol-anchored lipid protein transfer 1 |
-0.323 |
0.038 |
| 164 |
261199_at |
AT1G12950
|
RSH2, root hair specific 2 |
-0.322 |
-0.271 |
| 165 |
257131_at |
AT3G20240
|
[Mitochondrial substrate carrier family protein] |
-0.320 |
0.048 |
| 166 |
261228_at |
AT1G20050
|
HYD1, HYDRA1, MAD4, miRNA action deficient 4 |
-0.317 |
0.045 |
| 167 |
255516_at |
AT4G02270
|
RHS13, root hair specific 13 |
-0.317 |
-0.062 |
| 168 |
246015_at |
AT5G10700
|
[Peptidyl-tRNA hydrolase II (PTH2) family protein] |
-0.315 |
0.118 |
| 169 |
264577_at |
AT1G05260
|
RCI3, RARE COLD INDUCIBLE GENE 3, RCI3A |
-0.313 |
-0.119 |
| 170 |
251446_at |
AT3G59840
|
unknown |
-0.313 |
0.053 |
| 171 |
250992_at |
AT5G02260
|
ATEXP9, ATEXPA9, expansin A9, ATHEXP ALPHA 1.10, EXP9, EXPA9, expansin A9 |
-0.312 |
-0.032 |
| 172 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
-0.312 |
-0.251 |
| 173 |
247947_at |
AT5G57090
|
AGR, AGRAVITROPIC ROOT, AGR1, AGRAVITROPIC ROOT 1, ATPIN2, ARABIDOPSIS THALIANA PIN-FORMED 2, EIR1, ETHYLENE INSENSITIVE ROOT 1, MM31, PIN2, PIN-FORMED 2, WAV6, WAVY ROOTS 6 |
-0.309 |
-0.008 |
| 174 |
253291_at |
AT4G33865
|
[Ribosomal protein S14p/S29e family protein] |
-0.309 |
0.036 |
| 175 |
246403_at |
AT1G57590
|
[Pectinacetylesterase family protein] |
-0.308 |
0.048 |
| 176 |
253731_at |
AT4G29260
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.308 |
0.102 |
| 177 |
248790_at |
AT5G47450
|
ATTIP2;3, ARABIDOPSIS THALIANA TONOPLAST INTRINSIC PROTEIN 2;3, DELTA-TIP3, DELTA-TONOPLAST INTRINSIC PROTEIN 3, TIP2;3, tonoplast intrinsic protein 2;3 |
-0.306 |
-0.085 |
| 178 |
263482_at |
AT2G03980
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.305 |
0.015 |
| 179 |
262329_at |
AT1G64090
|
RTNLB3, Reticulan like protein B3 |
-0.304 |
0.065 |
| 180 |
255569_at |
AT4G01320
|
ATSTE24, STE24 |
-0.304 |
0.098 |
| 181 |
246793_at |
AT5G27210
|
Cand8, candidate G-protein Coupled Receptor 8 |
-0.301 |
-0.005 |
| 182 |
253063_at |
AT4G37640
|
ACA2, calcium ATPase 2 |
-0.300 |
0.037 |
| 183 |
249847_at |
AT5G23210
|
SCPL34, serine carboxypeptidase-like 34 |
-0.298 |
0.031 |
| 184 |
253942_at |
AT4G27010
|
EMB2788, EMBRYO DEFECTIVE 2788 |
-0.296 |
-0.112 |
| 185 |
248753_at |
AT5G47630
|
mtACP3, mitochondrial acyl carrier protein 3 |
-0.294 |
0.029 |
| 186 |
253047_at |
AT4G37295
|
unknown |
-0.293 |
-0.069 |
| 187 |
259933_at |
AT1G34350
|
unknown |
-0.289 |
0.057 |