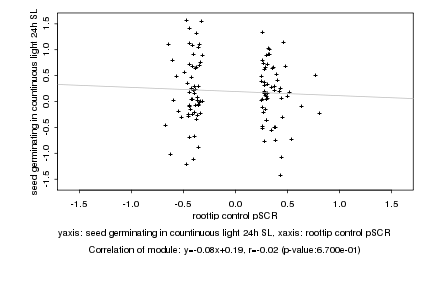
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
roottip control pSCR |
Log2 signal ratio
seed germinating in countinuous light 24h SL |
| 1 |
251044_at |
AT5G02350
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.799 |
-0.225 |
| 2 |
245845_at |
AT1G26150
|
AtPERK10, proline-rich extensin-like receptor kinase 10, PERK10, proline-rich extensin-like receptor kinase 10 |
0.762 |
0.504 |
| 3 |
262228_at |
AT1G68690
|
AtPERK9, proline-rich extensin-like receptor kinase 9, PERK9, proline-rich extensin-like receptor kinase 9 |
0.626 |
-0.085 |
| 4 |
249016_at |
AT5G44750
|
ATREV1, ARABIDOPSIS THALIANA HOMOLOG OF REVERSIONLESS 1, REV1 |
0.530 |
-0.715 |
| 5 |
265966_at |
AT2G37220
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.514 |
0.178 |
| 6 |
253519_at |
AT4G31240
|
AtNRX2, NRX2, nucleoredoxin 2 |
0.494 |
0.110 |
| 7 |
246653_at |
AT5G35200
|
[ENTH/ANTH/VHS superfamily protein] |
0.472 |
0.693 |
| 8 |
254724_at |
AT4G13630
|
unknown |
0.461 |
1.142 |
| 9 |
245702_at |
AT5G04220
|
ATSYTC, NTMC2T1.3, NTMC2TYPE1.3, SYT3, synaptotagmin 3, SYTC |
0.450 |
-0.296 |
| 10 |
245117_at |
AT2G41560
|
ACA4, autoinhibited Ca(2+)-ATPase, isoform 4 |
0.442 |
0.075 |
| 11 |
264223_s_at |
AT3G16030
|
CES101, CALLUS EXPRESSION OF RBCS 101, RFO3, RESISTANCE TO FUSARIUM OXYSPORUM 3 |
0.435 |
-1.079 |
| 12 |
247015_at |
AT5G66960
|
[Prolyl oligopeptidase family protein] |
0.431 |
0.259 |
| 13 |
266507_at |
AT2G47860
|
SETH6 |
0.426 |
-1.423 |
| 14 |
259429_at |
AT1G01600
|
CYP86A4, cytochrome P450, family 86, subfamily A, polypeptide 4 |
0.416 |
0.195 |
| 15 |
252962_at |
AT4G38780
|
[Pre-mRNA-processing-splicing factor] |
0.396 |
0.413 |
| 16 |
258255_at |
AT3G26800
|
unknown |
0.387 |
0.529 |
| 17 |
266733_at |
AT2G03280
|
[O-fucosyltransferase family protein] |
0.385 |
-0.483 |
| 18 |
245079_at |
AT2G23330
|
[copia-like retrotransposon family, has a 3.9e-195 P-value blast match to GB:AAA57005 Hopscotch polyprotein (Ty1_Copia-element) (Zea mays)] |
0.381 |
-0.747 |
| 19 |
248553_at |
AT5G50170
|
[C2 calcium/lipid-binding and GRAM domain containing protein] |
0.373 |
-0.483 |
| 20 |
253278_at |
AT4G34220
|
[Leucine-rich repeat protein kinase family protein] |
0.367 |
0.298 |
| 21 |
267015_at |
AT2G39340
|
AtSAC3A, SAC3A, yeast Sac3 homolog A |
0.366 |
0.228 |
| 22 |
255265_at |
AT4G05190
|
ATK5, kinesin 5 |
0.364 |
0.658 |
| 23 |
256661_at |
AT3G11964
|
RRP5, Ribosomal RNA Processing 5 |
0.347 |
0.640 |
| 24 |
263196_at |
AT1G36070
|
[Transducin/WD40 repeat-like superfamily protein] |
0.345 |
0.289 |
| 25 |
252820_at |
AT3G42640
|
AHA8, H(+)-ATPase 8, HA8, H(+)-ATPase 8 |
0.341 |
-0.556 |
| 26 |
251787_at |
AT3G55410
|
[2-oxoglutarate dehydrogenase, E1 component] |
0.323 |
1.016 |
| 27 |
264513_at |
AT1G09420
|
G6PD4, glucose-6-phosphate dehydrogenase 4 |
0.323 |
0.918 |
| 28 |
262068_at |
AT1G80070
|
EMB14, EMBRYO DEFECTIVE 14, EMB177, EMBRYO DEFECTIVE 177, EMB33, EMBRYO DEFECTIVE 33, SUS2, ABNORMAL SUSPENSOR 2 |
0.311 |
1.033 |
| 29 |
259681_at |
AT1G77760
|
GNR1, NIA1, nitrate reductase 1, NR1, NITRATE REDUCTASE 1 |
0.309 |
0.922 |
| 30 |
249550_at |
AT5G38210
|
[Protein kinase family protein] |
0.307 |
0.338 |
| 31 |
248547_at |
AT5G50280
|
EMB1006, embryo defective 1006 |
0.305 |
0.072 |
| 32 |
257963_at |
AT3G19840
|
ATPRP40C, pre-mRNA-processing protein 40C, PRP40C, pre-mRNA-processing protein 40C |
0.303 |
0.166 |
| 33 |
259422_at |
AT1G13810
|
[Restriction endonuclease, type II-like superfamily protein] |
0.302 |
0.718 |
| 34 |
260660_at |
AT1G19485
|
[Transducin/WD40 repeat-like superfamily protein] |
0.294 |
0.040 |
| 35 |
259987_at |
AT1G75030
|
ATLP-3, thaumatin-like protein 3, TLP-3, thaumatin-like protein 3 |
0.294 |
-0.354 |
| 36 |
266006_at |
AT2G37360
|
ABCG2, ATP-binding cassette G2 |
0.292 |
-0.364 |
| 37 |
266951_at |
AT2G18940
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.291 |
0.098 |
| 38 |
251897_at |
AT3G54360
|
NCA1, NO CATALASE ACTIVITY 1 |
0.289 |
0.894 |
| 39 |
266349_at |
AT2G01460
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.286 |
-0.150 |
| 40 |
261063_at |
AT1G07520
|
[GRAS family transcription factor] |
0.281 |
0.128 |
| 41 |
246897_at |
AT5G25560
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
0.280 |
0.668 |
| 42 |
247048_at |
AT5G66560
|
[Phototropic-responsive NPH3 family protein] |
0.278 |
0.198 |
| 43 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.277 |
-0.763 |
| 44 |
259903_at |
AT1G74160
|
TRM4, TON1 Recruiting Motif 4 |
0.277 |
0.180 |
| 45 |
254602_at |
AT4G19040
|
EDR2, ENHANCED DISEASE RESISTANCE 2 |
0.276 |
0.376 |
| 46 |
262299_at |
AT1G27520
|
[Glycosyl hydrolase family 47 protein] |
0.276 |
0.620 |
| 47 |
262109_at |
AT1G02730
|
ATCSLD5, cellulose synthase-like D5, CSLD5, CELLULOSE SYNTHASE LIKE D5, CSLD5, cellulose synthase-like D5, SOS6, SALT OVERLY SENSITIVE 6 |
0.274 |
0.156 |
| 48 |
262296_at |
AT1G27630
|
CYCT1;3, cyclin T 1;3 |
0.273 |
0.314 |
| 49 |
267406_at |
AT2G34780
|
EMB1611, EMBRYO DEFECTIVE 1611, MEE22, MATERNAL EFFECT EMBRYO ARREST 22 |
0.269 |
0.736 |
| 50 |
255899_at |
AT1G17970
|
[RING/U-box superfamily protein] |
0.266 |
-0.211 |
| 51 |
255639_at |
AT4G00760
|
APRR8, pseudo-response regulator 8, PRR8, PSEUDO-RESPONSE REGULATOR 8 |
0.258 |
0.800 |
| 52 |
248183_at |
AT5G54040
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.258 |
0.044 |
| 53 |
261979_at |
AT1G37130
|
ATNR2, ARABIDOPSIS NITRATE REDUCTASE 2, B29, CHL3, CHLORATE RESISTANT 3, NIA2, nitrate reductase 2, NIA2-1, NR, NITRATE REDUCTASE, NR2, NITRATE REDUCTASE 2 |
0.255 |
1.348 |
| 54 |
265302_at |
AT2G13970
|
[Mutator-like transposase family, has a 4.1e-39 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
0.254 |
-0.476 |
| 55 |
259036_at |
AT3G09220
|
LAC7, laccase 7 |
0.254 |
-0.110 |
| 56 |
262548_at |
AT1G31280
|
AGO2, argonaute 2, AtAGO2 |
0.251 |
-0.519 |
| 57 |
256930_at |
AT3G22460
|
OASA2, O-acetylserine (thiol) lyase (OAS-TL) isoform A2 |
0.248 |
0.396 |
| 58 |
265092_at |
AT1G03910
|
CTN, CACTIN |
0.248 |
0.487 |
| 59 |
259235_at |
AT3G11600
|
unknown |
0.246 |
0.023 |
| 60 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
-0.676 |
-0.455 |
| 61 |
250562_at |
AT5G08040
|
TOM5, mitochondrial import receptor subunit TOM5 homolog |
-0.648 |
1.119 |
| 62 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
-0.631 |
-1.005 |
| 63 |
251240_at |
AT3G62450
|
unknown |
-0.609 |
0.804 |
| 64 |
247882_at |
AT5G57785
|
unknown |
-0.604 |
0.028 |
| 65 |
252169_at |
AT3G50520
|
[Phosphoglycerate mutase family protein] |
-0.575 |
0.495 |
| 66 |
256563_at |
AT3G29780
|
RALFL27, ralf-like 27 |
-0.551 |
-0.177 |
| 67 |
262759_at |
AT1G10800
|
unknown |
-0.522 |
-0.292 |
| 68 |
249134_at |
AT5G43150
|
unknown |
-0.492 |
0.564 |
| 69 |
259195_at |
AT3G01730
|
unknown |
-0.478 |
-1.198 |
| 70 |
248972_at |
AT5G45010
|
ATDSS1(V), DSS1 homolog on chromosome V, DSS1(V), DSS1 homolog on chromosome V |
-0.477 |
1.583 |
| 71 |
251964_at |
AT3G53370
|
[S1FA-like DNA-binding protein] |
-0.465 |
0.353 |
| 72 |
246270_at |
AT4G36500
|
unknown |
-0.456 |
-0.236 |
| 73 |
245021_at |
ATCG00550
|
PSBJ, photosystem II reaction center protein J |
-0.455 |
-0.281 |
| 74 |
251355_at |
AT3G61100
|
[Putative endonuclease or glycosyl hydrolase] |
-0.451 |
0.183 |
| 75 |
251517_at |
AT3G59370
|
[Vacuolar calcium-binding protein-related] |
-0.450 |
-0.681 |
| 76 |
245495_at |
AT4G16400
|
unknown |
-0.448 |
-0.092 |
| 77 |
253291_at |
AT4G33865
|
[Ribosomal protein S14p/S29e family protein] |
-0.444 |
0.732 |
| 78 |
267084_at |
AT2G41180
|
SIB2, sigma factor binding protein 2 |
-0.444 |
-0.066 |
| 79 |
265883_at |
AT2G42310
|
unknown |
-0.442 |
1.413 |
| 80 |
250800_at |
AT5G05370
|
[Cytochrome b-c1 complex, subunit 8 protein] |
-0.442 |
1.139 |
| 81 |
250454_at |
AT5G09830
|
BolA2, homolog of E.coli BolA 2 |
-0.437 |
-0.144 |
| 82 |
260549_at |
AT2G43535
|
[Scorpion toxin-like knottin superfamily protein] |
-0.428 |
0.047 |
| 83 |
247006_at |
AT5G67490
|
unknown |
-0.427 |
0.270 |
| 84 |
253636_at |
AT4G30500
|
unknown |
-0.425 |
0.468 |
| 85 |
252390_at |
AT3G47836
|
unknown |
-0.415 |
0.688 |
| 86 |
262876_at |
AT1G64750
|
ATDSS1(I), deletion of SUV3 suppressor 1(I), DSS1(I), deletion of SUV3 suppressor 1(I) |
-0.414 |
1.088 |
| 87 |
263673_at |
AT2G04800
|
unknown |
-0.413 |
-0.241 |
| 88 |
253629_at |
AT4G30450
|
[glycine-rich protein] |
-0.413 |
0.044 |
| 89 |
246479_at |
AT5G16060
|
[Cytochrome c oxidase biogenesis protein Cmc1-like] |
-0.410 |
0.913 |
| 90 |
255074_at |
AT4G09100
|
[RING/U-box superfamily protein] |
-0.410 |
-1.119 |
| 91 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
-0.407 |
0.215 |
| 92 |
250923_at |
AT5G03455
|
ACR2, ARSENATE REDUCTASE 2, ARATH;CDC25, AtACR2, CDC25 |
-0.403 |
0.294 |
| 93 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.397 |
-0.667 |
| 94 |
246582_at |
AT1G31750
|
[proline-rich family protein] |
-0.394 |
0.157 |
| 95 |
260683_at |
AT1G17560
|
HLL, HUELLENLOS |
-0.394 |
-0.202 |
| 96 |
253482_at |
AT4G31985
|
[Ribosomal protein L39 family protein] |
-0.392 |
0.654 |
| 97 |
260867_at |
AT1G43790
|
TED6, tracheary element differentiation-related 6 |
-0.389 |
-0.058 |
| 98 |
263785_at |
AT2G46390
|
SDH8, succinate dehydrogenase 8 |
-0.387 |
0.246 |
| 99 |
249101_at |
AT5G43580
|
UPI, UNUSUAL SERINE PROTEASE INHIBITOR |
-0.379 |
-0.345 |
| 100 |
262558_at |
AT1G31335
|
unknown |
-0.379 |
0.676 |
| 101 |
266566_at |
AT2G24040
|
[Low temperature and salt responsive protein family] |
-0.376 |
1.324 |
| 102 |
248631_at |
AT5G49000
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.374 |
0.031 |
| 103 |
248208_at |
AT5G53980
|
ATHB52, homeobox protein 52, HB52, homeobox protein 52 |
-0.373 |
-0.260 |
| 104 |
253582_at |
AT4G30670
|
[Putative membrane lipoprotein] |
-0.366 |
0.078 |
| 105 |
258821_at |
AT3G07230
|
[wound-responsive protein-related] |
-0.364 |
1.044 |
| 106 |
255525_at |
AT4G02340
|
[alpha/beta-Hydrolases superfamily protein] |
-0.363 |
-0.073 |
| 107 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.361 |
-0.052 |
| 108 |
252502_at |
AT3G46900
|
COPT2, copper transporter 2 |
-0.358 |
-0.880 |
| 109 |
248746_at |
AT5G47890
|
[NADH-ubiquinone oxidoreductase B8 subunit, putative] |
-0.357 |
0.300 |
| 110 |
266796_at |
AT2G02880
|
[mucin-related] |
-0.353 |
1.117 |
| 111 |
255759_at |
AT1G16790
|
[ribosomal protein-related] |
-0.351 |
-0.003 |
| 112 |
260564_at |
AT2G43810
|
LSM6B, SM-like 6B |
-0.346 |
0.709 |
| 113 |
245334_at |
AT4G15800
|
RALFL33, ralf-like 33 |
-0.339 |
0.766 |
| 114 |
249863_at |
AT5G22950
|
VPS24.1 |
-0.338 |
-0.216 |
| 115 |
267178_at |
AT2G37750
|
unknown |
-0.336 |
0.005 |
| 116 |
258713_at |
AT3G09735
|
[S1FA-like DNA-binding protein] |
-0.333 |
1.548 |
| 117 |
256309_at |
AT1G30380
|
PSAK, photosystem I subunit K |
-0.324 |
0.893 |
| 118 |
247520_at |
AT5G61310
|
[Cytochrome c oxidase subunit Vc family protein] |
-0.324 |
0.007 |