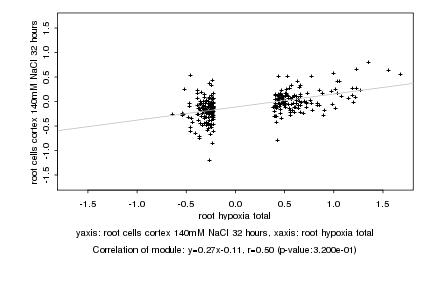
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
root hypoxia total |
Log2 signal ratio
root cells cortex 140mM NaCl 32 hours |
| 1 |
250464_at |
AT5G10040
|
unknown |
1.674 |
0.554 |
| 2 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
1.555 |
0.639 |
| 3 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
1.345 |
0.812 |
| 4 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
1.264 |
0.228 |
| 5 |
260668_at |
AT1G19530
|
unknown |
1.229 |
0.657 |
| 6 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
1.221 |
0.276 |
| 7 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
1.213 |
0.085 |
| 8 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
1.194 |
-0.015 |
| 9 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
1.189 |
0.135 |
| 10 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
1.187 |
0.269 |
| 11 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
1.142 |
0.066 |
| 12 |
254200_at |
AT4G24110
|
unknown |
1.072 |
0.122 |
| 13 |
257153_at |
AT3G27220
|
[Galactose oxidase/kelch repeat superfamily protein] |
1.051 |
0.423 |
| 14 |
250152_at |
AT5G15120
|
unknown |
1.035 |
0.413 |
| 15 |
257925_at |
AT3G23170
|
unknown |
1.029 |
0.180 |
| 16 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
1.012 |
-0.127 |
| 17 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
1.008 |
0.255 |
| 18 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
0.989 |
0.585 |
| 19 |
260522_x_at |
AT2G41730
|
unknown |
0.980 |
-0.050 |
| 20 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.968 |
0.218 |
| 21 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
0.904 |
-0.168 |
| 22 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
0.893 |
-0.278 |
| 23 |
249384_at |
AT5G39890
|
unknown |
0.879 |
0.181 |
| 24 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.853 |
-0.077 |
| 25 |
263295_at |
AT2G14210
|
AGL44, AGAMOUS-like 44, ANR1, ARABIDOPSIS NITRATE REGULATED 1 |
0.846 |
0.244 |
| 26 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.834 |
-0.068 |
| 27 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
0.827 |
-0.022 |
| 28 |
257922_at |
AT3G23150
|
ETR2, ethylene response 2 |
0.775 |
-0.164 |
| 29 |
261567_at |
AT1G33055
|
unknown |
0.771 |
0.511 |
| 30 |
245226_at |
AT3G29970
|
[B12D protein] |
0.769 |
-0.020 |
| 31 |
260451_at |
AT1G72360
|
AtERF73, ERF73, ethylene response factor 73, HRE1, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 1 |
0.757 |
0.031 |
| 32 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
0.728 |
0.181 |
| 33 |
259921_at |
AT1G72540
|
[Protein kinase superfamily protein] |
0.717 |
-0.035 |
| 34 |
251026_at |
AT5G02200
|
FHL, far-red-elongated hypocotyl1-like |
0.716 |
-0.010 |
| 35 |
245076_at |
AT2G23170
|
GH3.3 |
0.714 |
-0.111 |
| 36 |
253616_at |
AT4G30380
|
[Barwin-related endoglucanase] |
0.693 |
0.009 |
| 37 |
262383_at |
AT1G72940
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.684 |
-0.238 |
| 38 |
250472_at |
AT5G10210
|
unknown |
0.669 |
-0.012 |
| 39 |
267230_at |
AT2G44080
|
ARL, ARGOS-like |
0.660 |
0.146 |
| 40 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
0.657 |
0.118 |
| 41 |
249015_at |
AT5G44730
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.656 |
0.334 |
| 42 |
246200_at |
AT4G37240
|
unknown |
0.655 |
-0.216 |
| 43 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.653 |
-0.067 |
| 44 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.651 |
0.302 |
| 45 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.643 |
0.003 |
| 46 |
257163_at |
AT3G24310
|
ATMYB71, MYB DOMAIN PROTEIN 71, MYB305, myb domain protein 305 |
0.641 |
-0.063 |
| 47 |
252882_at |
AT4G39675
|
unknown |
0.637 |
-0.128 |
| 48 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
0.636 |
0.077 |
| 49 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
0.629 |
0.418 |
| 50 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
0.619 |
-0.020 |
| 51 |
253722_at |
AT4G29190
|
AtC3H49, AtOZF2, Arabidopsis thaliana oxidation-related zinc finger 2, AtTZF3, OZF2, oxidation-related zinc finger 2, TZF3, tandem zinc finger 3 |
0.618 |
0.103 |
| 52 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.614 |
0.026 |
| 53 |
245586_at |
AT4G14980
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.604 |
-0.109 |
| 54 |
245401_at |
AT4G17670
|
unknown |
0.599 |
-0.215 |
| 55 |
259685_at |
AT1G63090
|
AtPP2-A11, phloem protein 2-A11, PP2-A11, phloem protein 2-A11 |
0.599 |
-0.121 |
| 56 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
0.595 |
0.113 |
| 57 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.593 |
0.140 |
| 58 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.581 |
-0.029 |
| 59 |
253322_at |
AT4G33980
|
unknown |
0.579 |
0.089 |
| 60 |
253573_at |
AT4G31020
|
[alpha/beta-Hydrolases superfamily protein] |
0.579 |
-0.164 |
| 61 |
267083_at |
AT2G41100
|
ATCAL4, ARABIDOPSIS THALIANA CALMODULIN LIKE 4, TCH3, TOUCH 3 |
0.567 |
0.055 |
| 62 |
245998_at |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
0.567 |
0.332 |
| 63 |
266998_at |
AT2G34400
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.563 |
-0.049 |
| 64 |
256366_at |
AT1G66880
|
[Protein kinase superfamily protein] |
0.556 |
-0.183 |
| 65 |
262741_at |
AT1G28760
|
[Uncharacterized conserved protein (DUF2215)] |
0.556 |
0.062 |
| 66 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
0.550 |
-0.218 |
| 67 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
0.550 |
-0.278 |
| 68 |
249234_at |
AT5G42200
|
[RING/U-box superfamily protein] |
0.548 |
-0.129 |
| 69 |
251012_at |
AT5G02580
|
unknown |
0.543 |
0.282 |
| 70 |
252168_at |
AT3G50440
|
ATMES10, ARABIDOPSIS THALIANA METHYL ESTERASE 10, MES10, methyl esterase 10 |
0.540 |
0.266 |
| 71 |
264837_at |
AT1G03600
|
PSB27 |
0.535 |
0.072 |
| 72 |
264836_at |
AT1G03610
|
unknown |
0.535 |
0.109 |
| 73 |
259293_at |
AT3G11580
|
[AP2/B3-like transcriptional factor family protein] |
0.534 |
-0.044 |
| 74 |
258879_at |
AT3G03270
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.526 |
0.143 |
| 75 |
255945_at |
AT5G28610
|
unknown |
0.524 |
0.511 |
| 76 |
253987_at |
AT4G26270
|
PFK3, phosphofructokinase 3 |
0.514 |
0.143 |
| 77 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
0.514 |
0.171 |
| 78 |
266016_at |
AT2G18670
|
[RING/U-box superfamily protein] |
0.513 |
0.260 |
| 79 |
253264_at |
AT4G33950
|
ATOST1, OPEN STOMATA 1, OST1, OPEN STOMATA 1, P44, SNRK2-6, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-6, SNRK2.6, SNF1-RELATED PROTEIN KINASE 2.6, SRK2E |
0.508 |
-0.034 |
| 80 |
264561_at |
AT1G55810
|
UKL3, uridine kinase-like 3 |
0.507 |
0.113 |
| 81 |
263668_at |
AT1G04350
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.507 |
0.014 |
| 82 |
247024_at |
AT5G66985
|
unknown |
0.503 |
-0.111 |
| 83 |
261261_at |
AT1G26730
|
[EXS (ERD1/XPR1/SYG1) family protein] |
0.500 |
0.172 |
| 84 |
263931_at |
AT2G36220
|
unknown |
0.499 |
0.006 |
| 85 |
259979_at |
AT1G76600
|
unknown |
0.498 |
-0.079 |
| 86 |
245171_at |
AT2G47560
|
[RING/U-box superfamily protein] |
0.490 |
-0.057 |
| 87 |
263850_at |
AT2G04480
|
unknown |
0.487 |
0.008 |
| 88 |
260472_at |
AT1G10990
|
unknown |
0.484 |
-0.003 |
| 89 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
0.483 |
0.007 |
| 90 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
0.478 |
0.069 |
| 91 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
0.477 |
0.111 |
| 92 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
0.475 |
0.058 |
| 93 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
0.473 |
0.094 |
| 94 |
253404_at |
AT4G32840
|
PFK6, phosphofructokinase 6 |
0.469 |
0.123 |
| 95 |
251544_at |
AT3G58790
|
GAUT15, galacturonosyltransferase 15 |
0.469 |
0.108 |
| 96 |
258608_at |
AT3G03020
|
unknown |
0.468 |
0.228 |
| 97 |
260005_at |
AT1G67920
|
unknown |
0.466 |
0.143 |
| 98 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
0.466 |
0.066 |
| 99 |
248138_at |
AT5G54960
|
PDC2, pyruvate decarboxylase-2 |
0.460 |
-0.027 |
| 100 |
257466_at |
AT1G62840
|
unknown |
0.460 |
-0.038 |
| 101 |
260662_at |
AT1G19540
|
[NmrA-like negative transcriptional regulator family protein] |
0.459 |
-0.329 |
| 102 |
248404_at |
AT5G51460
|
ATTPPA, TPPA, trehalose-6-phosphate phosphatase A |
0.456 |
-0.146 |
| 103 |
245547_at |
AT4G15300
|
CYP702A2, cytochrome P450, family 702, subfamily A, polypeptide 2 |
0.454 |
0.176 |
| 104 |
257140_at |
AT3G28910
|
ATMYB30, MYB30, myb domain protein 30 |
0.452 |
-0.226 |
| 105 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
0.448 |
-0.056 |
| 106 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.446 |
-0.095 |
| 107 |
267384_at |
AT2G44370
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.445 |
-0.021 |
| 108 |
248160_at |
AT5G54470
|
BBX29, B-box domain protein 29 |
0.444 |
0.083 |
| 109 |
259996_at |
AT1G67910
|
unknown |
0.436 |
0.142 |
| 110 |
255742_at |
AT1G25560
|
AtTEM1, EDF1, ETHYLENE RESPONSE DNA BINDING FACTOR 1, TEM1, TEMPRANILLO 1 |
0.435 |
0.029 |
| 111 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
0.435 |
0.059 |
| 112 |
254667_at |
AT4G18280
|
[glycine-rich cell wall protein-related] |
0.434 |
0.158 |
| 113 |
267276_at |
AT2G30130
|
ASL5, LBD12, PCK1, PEACOCK 1 |
0.434 |
0.118 |
| 114 |
248719_at |
AT5G47910
|
ATRBOHD, RBOHD, respiratory burst oxidase homologue D |
0.432 |
0.064 |
| 115 |
253737_at |
AT4G28703
|
[RmlC-like cupins superfamily protein] |
0.432 |
0.525 |
| 116 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.430 |
0.070 |
| 117 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.429 |
0.070 |
| 118 |
259616_at |
AT1G47960
|
ATC/VIF1, CELL WALL / VACUOLAR INHIBITOR OF FRUCTOSIDASE 1, C/VIF1, cell wall / vacuolar inhibitor of fructosidase 1 |
0.428 |
-0.029 |
| 119 |
245324_at |
AT4G17260
|
[Lactate/malate dehydrogenase family protein] |
0.426 |
-0.100 |
| 120 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
0.419 |
-0.791 |
| 121 |
257766_at |
AT3G23030
|
IAA2, indole-3-acetic acid inducible 2 |
0.417 |
-0.122 |
| 122 |
266965_at |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
0.415 |
-0.426 |
| 123 |
261581_at |
AT1G01140
|
CIPK9, CBL-interacting protein kinase 9, PKS6, PROTEIN KINASE 6, SnRK3.12, SNF1-RELATED PROTEIN KINASE 3.12 |
0.412 |
-0.085 |
| 124 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
0.412 |
-0.132 |
| 125 |
255532_at |
AT4G02170
|
unknown |
0.411 |
-0.287 |
| 126 |
258493_at |
AT3G02555
|
unknown |
0.407 |
0.043 |
| 127 |
254522_at |
AT4G19980
|
unknown |
0.406 |
0.139 |
| 128 |
256262_at |
AT3G12150
|
unknown |
0.406 |
-0.105 |
| 129 |
265075_at |
AT1G55450
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.404 |
0.042 |
| 130 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
0.404 |
0.159 |
| 131 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
0.402 |
-0.024 |
| 132 |
265481_at |
AT2G15960
|
unknown |
0.400 |
-0.097 |
| 133 |
264647_at |
AT1G09090
|
ATRBOHB, respiratory burst oxidase homolog B, ATRBOHB-BETA, RBOHB, respiratory burst oxidase homolog B |
0.399 |
-0.287 |
| 134 |
248428_at |
AT5G51760
|
AHG1, ABA-hypersensitive germination 1 |
0.392 |
-0.184 |
| 135 |
247540_at |
AT5G61590
|
[Integrase-type DNA-binding superfamily protein] |
0.391 |
-0.294 |
| 136 |
257536_at |
AT3G02800
|
AtPFA-DSP3, PFA-DSP3, plant and fungi atypical dual-specificity phosphatase 3 |
0.391 |
-0.095 |
| 137 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.384 |
-0.111 |
| 138 |
267028_at |
AT2G38470
|
ATWRKY33, WRKY DNA-BINDING PROTEIN 33, WRKY33, WRKY DNA-binding protein 33 |
0.383 |
-0.106 |
| 139 |
254056_at |
AT4G25250
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.641 |
-0.255 |
| 140 |
247914_at |
AT5G57540
|
AtXTH13, XTH13, xyloglucan endotransglucosylase/hydrolase 13 |
-0.546 |
-0.271 |
| 141 |
253763_at |
AT4G28850
|
ATXTH26, XTH26, xyloglucan endotransglucosylase/hydrolase 26 |
-0.539 |
-0.243 |
| 142 |
258765_at |
AT3G10710
|
RHS12, root hair specific 12 |
-0.534 |
-0.253 |
| 143 |
264892_at |
AT1G23160
|
[Auxin-responsive GH3 family protein] |
-0.527 |
0.263 |
| 144 |
267222_at |
AT2G43880
|
[Pectin lyase-like superfamily protein] |
-0.484 |
-0.314 |
| 145 |
262864_at |
AT1G64920
|
[UDP-Glycosyltransferase superfamily protein] |
-0.472 |
-0.085 |
| 146 |
266222_at |
AT2G28780
|
unknown |
-0.471 |
-0.024 |
| 147 |
245571_at |
AT4G14695
|
[Uncharacterised protein family (UPF0041)] |
-0.461 |
0.551 |
| 148 |
258832_at |
AT3G07070
|
[Protein kinase superfamily protein] |
-0.460 |
-0.511 |
| 149 |
267217_at |
AT2G02610
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.457 |
-0.601 |
| 150 |
255591_at |
AT4G01630
|
ATEXP17, EXPANSIN 17, ATEXPA17, expansin A17, ATHEXP ALPHA 1.13, EXPA17, expansin A17 |
-0.452 |
-0.329 |
| 151 |
254632_at |
AT4G18630
|
unknown |
-0.441 |
-0.415 |
| 152 |
261550_at |
AT1G63450
|
RHS8, root hair specific 8, XUT1, XYLOGLUCAN-SPECIFIC GALACTURONOSYLTRANSFERASE 1 |
-0.410 |
-0.650 |
| 153 |
251405_at |
AT3G60330
|
AHA7, H(+)-ATPase 7, HA7, H(+)-ATPase 7 |
-0.394 |
-0.262 |
| 154 |
263932_at |
AT2G35990
|
LOG2, LONELY GUY 2 |
-0.390 |
-0.063 |
| 155 |
248209_at |
AT5G53990
|
[UDP-Glycosyltransferase superfamily protein] |
-0.390 |
0.180 |
| 156 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
-0.389 |
0.238 |
| 157 |
248431_at |
AT5G51470
|
[Auxin-responsive GH3 family protein] |
-0.388 |
0.062 |
| 158 |
253734_at |
AT4G29180
|
RHS16, root hair specific 16 |
-0.383 |
-0.381 |
| 159 |
266828_at |
AT2G22930
|
[UDP-Glycosyltransferase superfamily protein] |
-0.377 |
-0.129 |
| 160 |
259982_at |
AT1G76410
|
ATL8 |
-0.376 |
-0.246 |
| 161 |
255608_at |
AT4G01140
|
unknown |
-0.373 |
-0.695 |
| 162 |
267137_at |
AT2G23410
|
ACPT, cis-prenyltransferase, AtcPT3, CPT, cis-prenyltransferase, cPT3, cis-prenyltransferase 3 |
-0.373 |
-0.752 |
| 163 |
263376_at |
AT2G20520
|
FLA6, FASCICLIN-like arabinogalactan 6 |
-0.371 |
-0.129 |
| 164 |
248916_at |
AT5G45840
|
[Leucine-rich repeat protein kinase family protein] |
-0.364 |
-0.437 |
| 165 |
258178_at |
AT3G21680
|
unknown |
-0.363 |
-0.182 |
| 166 |
266900_at |
AT2G34610
|
unknown |
-0.361 |
-0.033 |
| 167 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
-0.358 |
0.007 |
| 168 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
-0.357 |
-0.049 |
| 169 |
259520_at |
AT1G12320
|
unknown |
-0.350 |
-0.030 |
| 170 |
262275_at |
AT1G68710
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
-0.346 |
-0.299 |
| 171 |
266797_at |
AT2G22840
|
AtGRF1, growth-regulating factor 1, GRF1, growth-regulating factor 1 |
-0.346 |
0.189 |
| 172 |
247830_at |
AT5G58530
|
[Glutaredoxin family protein] |
-0.345 |
-0.125 |
| 173 |
262972_at |
AT1G75620
|
[glyoxal oxidase-related protein] |
-0.343 |
-0.479 |
| 174 |
256953_at |
AT3G29630
|
[UDP-Glycosyltransferase superfamily protein] |
-0.340 |
-0.236 |
| 175 |
247498_at |
AT5G61810
|
APC1, ATP/phosphate carrier 1 |
-0.337 |
-0.105 |
| 176 |
254110_at |
AT4G25260
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.333 |
0.004 |
| 177 |
256138_at |
AT1G48670
|
[auxin-responsive GH3 family protein] |
-0.330 |
-0.082 |
| 178 |
260366_at |
AT1G70460
|
AtPERK13, proline-rich extensin-like receptor kinase 13, PERK13, proline-rich extensin-like receptor kinase 13, RHS10, root hair specific 10 |
-0.330 |
-0.199 |
| 179 |
246229_at |
AT4G37160
|
sks15, SKU5 similar 15 |
-0.325 |
-0.074 |
| 180 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
-0.320 |
-0.145 |
| 181 |
252594_at |
AT3G45680
|
[Major facilitator superfamily protein] |
-0.317 |
0.145 |
| 182 |
251144_at |
AT5G01210
|
[HXXXD-type acyl-transferase family protein] |
-0.315 |
-0.334 |
| 183 |
263616_at |
AT2G04680
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.315 |
-0.447 |
| 184 |
261226_at |
AT1G20190
|
ATEXP11, ATEXPA11, expansin 11, ATHEXP ALPHA 1.14, EXP11, EXPANSIN 11, EXPA11, expansin 11 |
-0.315 |
0.100 |
| 185 |
254697_at |
AT4G17970
|
ALMT12, aluminum-activated, malate transporter 12, ATALMT12, QUAC1, quick-activating anion channel 1 |
-0.315 |
-0.298 |
| 186 |
252858_at |
AT4G39770
|
TPPH, trehalose-6-phosphate phosphatase H |
-0.313 |
-0.093 |
| 187 |
249765_at |
AT5G24030
|
SLAH3, SLAC1 homologue 3 |
-0.312 |
-0.156 |
| 188 |
259736_at |
AT1G64390
|
AtGH9C2, glycosyl hydrolase 9C2, GH9C2, glycosyl hydrolase 9C2 |
-0.308 |
-0.039 |
| 189 |
254515_at |
AT4G20270
|
BAM3, BARELY ANY MERISTEM 3 |
-0.308 |
-0.318 |
| 190 |
267550_at |
AT2G32800
|
AP4.3A, LecRK-S.2, L-type lectin receptor kinase S.2 |
-0.303 |
-0.220 |
| 191 |
257815_at |
AT3G25130
|
unknown |
-0.303 |
0.008 |
| 192 |
253165_at |
AT4G35320
|
unknown |
-0.298 |
-0.196 |
| 193 |
265962_at |
AT2G37460
|
UMAMIT12, Usually multiple acids move in and out Transporters 12 |
-0.298 |
-0.315 |
| 194 |
256759_at |
AT3G25640
|
unknown |
-0.298 |
-0.418 |
| 195 |
251191_at |
AT3G62590
|
[alpha/beta-Hydrolases superfamily protein] |
-0.298 |
0.033 |
| 196 |
256128_at |
AT1G18140
|
ATLAC1, LAC1, laccase 1 |
-0.298 |
-0.478 |
| 197 |
258676_at |
AT3G08600
|
unknown |
-0.296 |
-0.244 |
| 198 |
263112_at |
AT1G03080
|
NET1D, Networked 1D |
-0.296 |
-0.151 |
| 199 |
249309_at |
AT5G41410
|
BEL1, BELL 1 |
-0.294 |
-0.169 |
| 200 |
249868_at |
AT5G23030
|
TET12, tetraspanin12 |
-0.289 |
-0.575 |
| 201 |
261099_at |
AT1G62980
|
ATEXP18, ATEXPA18, expansin A18, ATHEXP ALPHA 1.25, EXP18, EXPANSIN 18, EXPA18, expansin A18 |
-0.288 |
-0.196 |
| 202 |
266191_at |
AT2G39040
|
[Peroxidase superfamily protein] |
-0.288 |
-0.168 |
| 203 |
249169_at |
AT5G42880
|
unknown |
-0.284 |
-0.122 |
| 204 |
249136_at |
AT5G43180
|
unknown |
-0.283 |
-0.432 |
| 205 |
249768_at |
AT5G24100
|
[Leucine-rich repeat protein kinase family protein] |
-0.280 |
-0.438 |
| 206 |
245245_at |
AT1G44318
|
hemb2 |
-0.280 |
-0.326 |
| 207 |
245546_at |
AT4G15290
|
ATCSLB05, ATCSLB5, CELLULOSE SYNTHASE LIKE 5, CSLB05 |
-0.280 |
-0.538 |
| 208 |
263319_at |
AT2G47160
|
AtBOR1, BOR1, REQUIRES HIGH BORON 1 |
-0.278 |
-0.099 |
| 209 |
250968_at |
AT5G02890
|
[HXXXD-type acyl-transferase family protein] |
-0.278 |
-0.449 |
| 210 |
246329_at |
AT3G43610
|
[Spc97 / Spc98 family of spindle pole body (SBP) component] |
-0.277 |
0.009 |
| 211 |
256176_at |
AT1G51640
|
ATEXO70G2, exocyst subunit exo70 family protein G2, EXO70G2, exocyst subunit exo70 family protein G2 |
-0.276 |
-0.022 |
| 212 |
266078_at |
AT2G40670
|
ARR16, response regulator 16, RR16, response regulator 16 |
-0.275 |
0.006 |
| 213 |
253786_at |
AT4G28650
|
[Leucine-rich repeat transmembrane protein kinase family protein] |
-0.274 |
-0.212 |
| 214 |
251048_at |
AT5G02410
|
ALG10, homolog of yeast ALG10 |
-0.273 |
-0.245 |
| 215 |
255829_at |
AT2G40540
|
ATKT2, ATKUP2, KT2, potassium transporter 2, KUP2, SHY3, TRK2 |
-0.273 |
-0.057 |
| 216 |
256811_at |
AT3G21340
|
[Leucine-rich repeat protein kinase family protein] |
-0.270 |
-1.204 |
| 217 |
263330_at |
AT2G15320
|
[Leucine-rich repeat (LRR) family protein] |
-0.270 |
-0.248 |
| 218 |
259525_at |
AT1G12560
|
ATEXP7, ATEXPA7, expansin A7, ATHEXP ALPHA 1.26, EXP7, EXPA7, expansin A7 |
-0.270 |
-0.253 |
| 219 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
-0.268 |
0.063 |
| 220 |
251476_at |
AT3G59670
|
unknown |
-0.266 |
0.383 |
| 221 |
254644_at |
AT4G18510
|
CLE2, CLAVATA3/ESR-related 2 |
-0.266 |
-0.381 |
| 222 |
252536_at |
AT3G45700
|
[Major facilitator superfamily protein] |
-0.265 |
-0.178 |
| 223 |
249113_at |
AT5G43790
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.264 |
0.097 |
| 224 |
246625_at |
AT1G48880
|
TBL7, TRICHOME BIREFRINGENCE-LIKE 7 |
-0.262 |
-0.048 |
| 225 |
254294_at |
AT4G23070
|
ATRBL7, RHOMBOID-like protein 7, RBL7, RHOMBOID-like protein 7 |
-0.261 |
-0.486 |
| 226 |
250744_at |
AT5G05840
|
unknown |
-0.258 |
-0.212 |
| 227 |
250370_at |
AT5G11430
|
[SPOC domain / Transcription elongation factor S-II protein] |
-0.255 |
-0.111 |
| 228 |
255732_at |
AT1G25450
|
CER60, ECERIFERUM 60, KCS5, 3-ketoacyl-CoA synthase 5 |
-0.255 |
-0.671 |
| 229 |
262780_at |
AT1G13090
|
CYP71B28, cytochrome P450, family 71, subfamily B, polypeptide 28 |
-0.253 |
-0.049 |
| 230 |
250802_at |
AT5G04970
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.252 |
-0.180 |
| 231 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
-0.251 |
-0.182 |
| 232 |
257113_at |
AT3G20130
|
CYP705A22, cytochrome P450, family 705, subfamily A, polypeptide 22, GPS1, gravity persistence signal 1 |
-0.251 |
-0.294 |
| 233 |
255111_at |
AT4G08780
|
[Peroxidase superfamily protein] |
-0.251 |
0.142 |
| 234 |
254109_at |
AT4G25240
|
SKS1, SKU5 similar 1 |
-0.251 |
-0.002 |
| 235 |
259763_at |
AT1G77630
|
LYM3, lysin-motif (LysM) domain protein 3, LYP3, LysM-containing receptor protein 3 |
-0.251 |
0.341 |
| 236 |
266625_at |
AT2G35380
|
[Peroxidase superfamily protein] |
-0.251 |
-0.071 |
| 237 |
245967_at |
AT5G19800
|
[hydroxyproline-rich glycoprotein family protein] |
-0.251 |
-0.376 |
| 238 |
259588_at |
AT1G27930
|
[FUNCTIONS IN: protein binding] |
-0.249 |
-0.040 |
| 239 |
247097_at |
AT5G66460
|
AtMAN7, MAN7, endo-beta-mannase 7 |
-0.249 |
-0.077 |
| 240 |
267372_at |
AT2G26290
|
ARSK1, root-specific kinase 1 |
-0.249 |
-0.467 |
| 241 |
258718_at |
AT3G09760
|
[RING/U-box superfamily protein] |
-0.247 |
-0.161 |
| 242 |
259832_at |
AT1G69580
|
[Homeodomain-like superfamily protein] |
-0.245 |
-0.510 |
| 243 |
251060_at |
AT5G01820
|
ATCIPK14, ATSR1, serine/threonine protein kinase 1, CIPK14, CBL-INTERACTING PROTEIN KINASE 14, PKS24, SOS2-like protein kinase 24, SnRK3.15, SNF1-RELATED PROTEIN KINASE 3.15, SR1, serine/threonine protein kinase 1 |
-0.245 |
-0.093 |
| 244 |
262406_at |
AT1G34670
|
AtMYB93, myb domain protein 93, MYB93, myb domain protein 93 |
-0.243 |
-0.168 |
| 245 |
247057_at |
AT5G66770
|
[GRAS family transcription factor] |
-0.243 |
-0.032 |
| 246 |
266962_at |
AT2G39435
|
TRM18, TON1 Recruiting Motif 18 |
-0.243 |
-0.207 |
| 247 |
252046_at |
AT3G52460
|
[hydroxyproline-rich glycoprotein family protein] |
-0.243 |
-0.316 |
| 248 |
264144_at |
AT1G79320
|
AtMC6, metacaspase 6, AtMCP2c, metacaspase 2c, MC6, metacaspase 6, MCP2c, metacaspase 2c |
-0.242 |
-0.297 |
| 249 |
250173_at |
AT5G14340
|
AtMYB40, myb domain protein 40, MYB40, myb domain protein 40 |
-0.242 |
-0.853 |
| 250 |
255622_at |
AT4G01070
|
GT72B1, UGT72B1, UDP-GLUCOSE-DEPENDENT GLUCOSYLTRANSFERASE 72 B1 |
-0.242 |
-0.323 |
| 251 |
262448_at |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.240 |
-0.216 |
| 252 |
255773_at |
AT1G18590
|
ATSOT17, SULFOTRANSFERASE 17, ATST5C, ARABIDOPSIS SULFOTRANSFERASE 5C, SOT17, sulfotransferase 17 |
-0.239 |
0.066 |
| 253 |
262396_at |
AT1G49470
|
unknown |
-0.239 |
-0.230 |
| 254 |
263241_at |
AT2G16500
|
ADC1, arginine decarboxylase 1, ARGDC, ARGDC1, SPE1 |
-0.238 |
-0.172 |
| 255 |
246591_at |
AT5G14880
|
KUP8, potassium uptake 8 |
-0.237 |
-0.249 |
| 256 |
260410_at |
AT1G69870
|
AtNPF2.13, NPF2.13, NRT1/ PTR family 2.13, NRT1.7, nitrate transporter 1.7 |
-0.237 |
0.054 |
| 257 |
248853_at |
AT5G46570
|
BSK2, brassinosteroid-signaling kinase 2 |
-0.236 |
-0.100 |
| 258 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
-0.236 |
0.437 |
| 259 |
248066_at |
AT5G55590
|
QRT1, QUARTET 1 |
-0.236 |
-0.150 |
| 260 |
266765_at |
AT2G46860
|
AtPPa3, pyrophosphorylase 3, PPa3, pyrophosphorylase 3 |
-0.233 |
-0.457 |
| 261 |
247027_at |
AT5G67090
|
[Subtilisin-like serine endopeptidase family protein] |
-0.233 |
-0.120 |
| 262 |
255175_at |
AT4G07960
|
ATCSLC12, Cellulose-synthase-like C12, CSLC12, CELLULOSE-SYNTHASE LIKE C12, CSLC12, Cellulose-synthase-like C12 |
-0.231 |
-0.361 |
| 263 |
253613_at |
AT4G30320
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.231 |
-0.202 |
| 264 |
266300_at |
AT2G01420
|
ATPIN4, ARABIDOPSIS PIN-FORMED 4, PIN4, PIN-FORMED 4 |
-0.230 |
-0.087 |
| 265 |
250065_at |
AT5G17910
|
unknown |
-0.230 |
-0.266 |
| 266 |
252272_at |
AT3G49670
|
BAM2, BARELY ANY MERISTEM 2 |
-0.229 |
-0.127 |
| 267 |
249934_at |
AT5G22410
|
RHS18, root hair specific 18 |
-0.229 |
-0.096 |
| 268 |
253533_at |
AT4G31590
|
ATCSLC05, CELLULOSE-SYNTHASE LIKE C5, ATCSLC5, Cellulose-synthase-like C5, CSLC05, CELLULOSE-SYNTHASE LIKE C5, CSLC5, Cellulose-synthase-like C5 |
-0.229 |
0.076 |
| 269 |
265873_at |
AT2G01630
|
[O-Glycosyl hydrolases family 17 protein] |
-0.229 |
0.164 |
| 270 |
256293_at |
AT1G69440
|
AGO7, ARGONAUTE7, ZIP, ZIPPY |
-0.228 |
-0.017 |
| 271 |
264896_at |
AT1G23210
|
AtGH9B6, glycosyl hydrolase 9B6, GH9B6, glycosyl hydrolase 9B6 |
-0.227 |
0.078 |
| 272 |
247533_at |
AT5G61570
|
[Protein kinase superfamily protein] |
-0.227 |
-0.598 |
| 273 |
246826_at |
AT5G26310
|
UGT72E3 |
-0.225 |
-0.311 |
| 274 |
250591_at |
AT5G07720
|
XXT3, xyloglucan xylosyltransferase 3 |
-0.225 |
-0.103 |
| 275 |
247284_at |
AT5G64410
|
ATOPT4, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 4, OPT4, oligopeptide transporter 4 |
-0.224 |
-0.029 |