|
probeID |
AGICode |
Annotation |
Log2 signal ratio
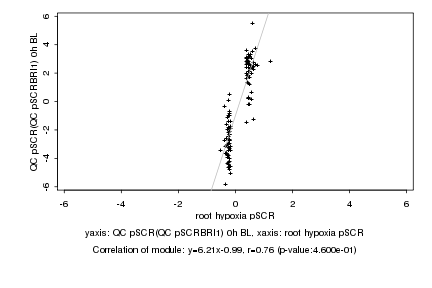
root hypoxia pSCR |
Log2 signal ratio
QC pSCR(QC pSCRBRI1) 0h BL |
| 1 |
266597_at |
AT2G46130
|
ATWRKY43, WRKY DNA-BINDING PROTEIN 43, WRKY43, WRKY DNA-binding protein 43 |
1.197 |
2.827 |
| 2 |
260535_at |
AT2G43390
|
unknown |
0.770 |
2.550 |
| 3 |
257704_at |
AT3G12720
|
ATMYB67, myb domain protein 67, ATY53, MYB67, MYB67, myb domain protein 67 |
0.693 |
3.750 |
| 4 |
262339_at |
AT1G64000
|
ATWRKY56, WRKY56, WRKY DNA-binding protein 56 |
0.674 |
2.642 |
| 5 |
260091_at |
AT1G73290
|
scpl5, serine carboxypeptidase-like 5 |
0.632 |
2.811 |
| 6 |
252195_at |
AT3G50190
|
unknown |
0.625 |
2.306 |
| 7 |
255903_at |
AT1G17950
|
ATMYB52, MYB DOMAIN PROTEIN 52, BW52, MYB52, myb domain protein 52 |
0.612 |
2.486 |
| 8 |
245845_at |
AT1G26150
|
AtPERK10, proline-rich extensin-like receptor kinase 10, PERK10, proline-rich extensin-like receptor kinase 10 |
0.599 |
-1.245 |
| 9 |
252988_at |
AT4G38410
|
[Dehydrin family protein] |
0.592 |
5.529 |
| 10 |
254404_at |
AT4G21340
|
B70 |
0.592 |
2.421 |
| 11 |
248990_at |
AT5G45210
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.589 |
3.539 |
| 12 |
256131_at |
AT1G13600
|
AtbZIP58, basic leucine-zipper 58, bZIP58, basic leucine-zipper 58 |
0.550 |
2.012 |
| 13 |
267391_at |
AT2G44480
|
BGLU17, beta glucosidase 17 |
0.545 |
3.087 |
| 14 |
264437_at |
AT1G27510
|
EX2, EXECUTER 2 |
0.543 |
0.172 |
| 15 |
260672_at |
AT1G19480
|
[DNA glycosylase superfamily protein] |
0.540 |
0.703 |
| 16 |
263387_at |
AT2G40160
|
TBL30 |
0.512 |
2.343 |
| 17 |
261385_at |
AT1G05450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.500 |
3.368 |
| 18 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
0.494 |
2.537 |
| 19 |
248424_at |
AT5G51680
|
[hydroxyproline-rich glycoprotein family protein] |
0.484 |
-0.157 |
| 20 |
245345_at |
AT4G16640
|
[Matrixin family protein] |
0.480 |
2.164 |
| 21 |
247231_at |
AT5G65230
|
AtMYB53, myb domain protein 53, MYB53, myb domain protein 53 |
0.478 |
3.224 |
| 22 |
252196_at |
AT3G50200
|
unknown |
0.471 |
1.761 |
| 23 |
247769_at |
AT5G58910
|
LAC16, laccase 16 |
0.469 |
2.664 |
| 24 |
263005_at |
AT1G54540
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.462 |
3.221 |
| 25 |
263467_at |
AT2G31730
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.461 |
2.609 |
| 26 |
256710_at |
AT3G30350
|
GLV3, GOLVEN 3, RGF4, root meristem growth factor 4 |
0.459 |
1.204 |
| 27 |
256176_at |
AT1G51640
|
ATEXO70G2, exocyst subunit exo70 family protein G2, EXO70G2, exocyst subunit exo70 family protein G2 |
0.451 |
2.640 |
| 28 |
263825_at |
AT2G40370
|
LAC5, laccase 5 |
0.449 |
3.321 |
| 29 |
250572_at |
AT5G08210
|
MIR834A, microRNA834A |
0.447 |
0.219 |
| 30 |
258625_at |
AT3G04370
|
PDLP4, plasmodesmata-located protein 4 |
0.443 |
3.134 |
| 31 |
263327_at |
AT2G15300
|
[Leucine-rich repeat protein kinase family protein] |
0.442 |
2.153 |
| 32 |
262640_at |
AT1G62760
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.438 |
3.143 |
| 33 |
249794_at |
AT5G23530
|
AtCXE18, carboxyesterase 18, CXE18, carboxyesterase 18 |
0.433 |
0.334 |
| 34 |
260684_at |
AT1G17590
|
NF-YA8, nuclear factor Y, subunit A8 |
0.432 |
0.265 |
| 35 |
265846_at |
AT2G35770
|
scpl28, serine carboxypeptidase-like 28 |
0.431 |
1.869 |
| 36 |
250474_at |
AT5G10230
|
ANN7, ANNEXIN 7, ANNAT7, annexin 7 |
0.430 |
2.872 |
| 37 |
254415_at |
AT4G21326
|
ATSBT3.12, subtilase 3.12, SBT3.12, subtilase 3.12 |
0.427 |
-0.161 |
| 38 |
267048_at |
AT2G34200
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.427 |
1.332 |
| 39 |
266126_at |
AT2G45040
|
[Matrixin family protein] |
0.426 |
2.414 |
| 40 |
246181_at |
AT5G20860
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.422 |
1.877 |
| 41 |
259624_at |
AT1G43020
|
unknown |
0.421 |
2.716 |
| 42 |
248205_at |
AT5G54300
|
unknown |
0.420 |
1.385 |
| 43 |
260035_at |
AT1G68850
|
[Peroxidase superfamily protein] |
0.401 |
2.905 |
| 44 |
264010_at |
AT2G21100
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.400 |
3.065 |
| 45 |
248761_at |
AT5G47635
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.397 |
2.806 |
| 46 |
253397_at |
AT4G32710
|
PERK14, proline-rich extensin-like receptor kinase 14 |
0.393 |
1.843 |
| 47 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
0.386 |
3.112 |
| 48 |
245702_at |
AT5G04220
|
ATSYTC, NTMC2T1.3, NTMC2TYPE1.3, SYT3, synaptotagmin 3, SYTC |
0.384 |
2.030 |
| 49 |
259282_at |
AT3G11430
|
ATGPAT5, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 5, GPAT5, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 5 |
0.373 |
3.074 |
| 50 |
252233_at |
AT3G49690
|
ATMYB84, MYB DOMAIN PROTEIN 84, MYB84, myb domain protein 84, RAX3, REGULATOR OF AXILLARY MERISTEMS3 |
0.371 |
2.655 |
| 51 |
254914_at |
AT4G11290
|
[Peroxidase superfamily protein] |
0.370 |
3.639 |
| 52 |
246826_at |
AT5G26310
|
UGT72E3 |
0.369 |
2.463 |
| 53 |
248696_at |
AT5G48360
|
[Actin-binding FH2 (formin homology 2) family protein] |
0.369 |
1.631 |
| 54 |
267344_at |
AT2G44230
|
unknown |
0.368 |
2.877 |
| 55 |
252569_at |
AT3G45420
|
LecRK-I.4, L-type lectin receptor kinase I.4 |
0.367 |
-1.446 |
| 56 |
250157_at |
AT5G15180
|
[Peroxidase superfamily protein] |
-0.545 |
-3.428 |
| 57 |
252430_at |
AT3G47470
|
CAB4, LHCA4, light-harvesting chlorophyll-protein complex I subunit A4 |
-0.408 |
-0.299 |
| 58 |
266649_at |
AT2G25810
|
TIP4;1, tonoplast intrinsic protein 4;1 |
-0.404 |
-2.674 |
| 59 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
-0.370 |
-3.128 |
| 60 |
265050_at |
AT1G52070
|
[Mannose-binding lectin superfamily protein] |
-0.349 |
-5.772 |
| 61 |
267191_at |
AT2G44110
|
ATMLO15, MILDEW RESISTANCE LOCUS O 15, MLO15, MILDEW RESISTANCE LOCUS O 15 |
-0.349 |
-3.612 |
| 62 |
248551_at |
AT5G50200
|
ATNRT3.1, NRT3.1, NITRATE TRANSPORTER 3.1, WR3, WOUND-RESPONSIVE 3 |
-0.341 |
-2.580 |
| 63 |
266669_at |
AT2G29750
|
UGT71C1, UDP-glucosyl transferase 71C1 |
-0.338 |
-3.058 |
| 64 |
264826_at |
AT1G03410
|
2A6 |
-0.337 |
-3.624 |
| 65 |
255016_at |
AT4G10120
|
ATSPS4F |
-0.334 |
-3.600 |
| 66 |
254107_at |
AT4G25220
|
AtG3Pp2, glycerol-3-phosphate permease 2, G3Pp2, glycerol-3-phosphate permease 2, RHS15, root hair specific 15 |
-0.332 |
-1.599 |
| 67 |
254041_at |
AT4G25830
|
[Uncharacterised protein family (UPF0497)] |
-0.310 |
-1.067 |
| 68 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
-0.307 |
-3.776 |
| 69 |
265463_at |
AT2G37090
|
IRX9, IRREGULAR XYLEM 9 |
-0.306 |
-3.885 |
| 70 |
248723_at |
AT5G47950
|
[HXXXD-type acyl-transferase family protein] |
-0.303 |
-3.691 |
| 71 |
259268_at |
AT3G01070
|
AtENODL16, ENODL16, early nodulin-like protein 16 |
-0.296 |
-2.964 |
| 72 |
265852_at |
AT2G42350
|
[RING/U-box superfamily protein] |
-0.289 |
-1.901 |
| 73 |
258183_at |
AT3G21550
|
AtDMP2, Arabidopsis thaliana DUF679 domain membrane protein 2, DMP2, DUF679 domain membrane protein 2 |
-0.284 |
-4.347 |
| 74 |
247483_at |
AT5G62420
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.276 |
-2.944 |
| 75 |
264574_at |
AT1G05300
|
ZIP5, zinc transporter 5 precursor |
-0.276 |
-2.657 |
| 76 |
259774_at |
AT1G29520
|
[AWPM-19-like family protein] |
-0.271 |
-4.611 |
| 77 |
266670_at |
AT2G29740
|
UGT71C2, UDP-glucosyl transferase 71C2 |
-0.266 |
-0.939 |
| 78 |
249868_at |
AT5G23030
|
TET12, tetraspanin12 |
-0.265 |
-2.157 |
| 79 |
248885_at |
AT5G46150
|
[LEM3 (ligand-effect modulator 3) family protein / CDC50 family protein] |
-0.262 |
0.087 |
| 80 |
262388_at |
AT1G49320
|
unknown |
-0.258 |
-3.781 |
| 81 |
253626_at |
AT4G30640
|
[RNI-like superfamily protein] |
-0.257 |
-1.798 |
| 82 |
267582_at |
AT2G41970
|
[Protein kinase superfamily protein] |
-0.255 |
-2.453 |
| 83 |
247970_at |
AT5G56720
|
c-NAD-MDH3, cytosolic-NAD-dependent malate dehydrogenase 3 |
-0.249 |
-4.429 |
| 84 |
265279_at |
AT2G28460
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.247 |
-4.285 |
| 85 |
252502_at |
AT3G46900
|
COPT2, copper transporter 2 |
-0.247 |
-3.398 |
| 86 |
246868_at |
AT5G26010
|
[Protein phosphatase 2C family protein] |
-0.245 |
-1.380 |
| 87 |
267128_at |
AT2G23620
|
ATMES1, ARABIDOPSIS THALIANA METHYL ESTERASE 1, MES1, methyl esterase 1 |
-0.245 |
-3.173 |
| 88 |
257896_at |
AT3G16920
|
ATCTL2, CTL2, chitinase-like protein 2 |
-0.237 |
-3.988 |
| 89 |
247882_at |
AT5G57785
|
unknown |
-0.233 |
-0.915 |
| 90 |
262525_at |
AT1G17060
|
CHI2, CHIBI 2, CYP72C1, cytochrome p450 72c1, SHK1, SHRINK 1, SOB7, SUPPRESSOR OF PHYB-4 7 |
-0.233 |
-2.257 |
| 91 |
251255_at |
AT3G62280
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.232 |
-2.698 |
| 92 |
248263_at |
AT5G53370
|
ATPMEPCRF, PECTIN METHYLESTERASE PCR FRAGMENT F, PMEPCRF, pectin methylesterase PCR fragment F |
-0.224 |
-2.061 |
| 93 |
260553_at |
AT2G41800
|
unknown |
-0.224 |
-4.517 |
| 94 |
247902_at |
AT5G57350
|
AHA3, H(+)-ATPase 3, ATAHA3, ARABIDOPSIS THALIANA ARABIDOPSIS H(+)-ATPASE, HA3, H(+)-ATPase 3 |
-0.223 |
-4.755 |
| 95 |
253816_at |
AT4G28210
|
EMB1923, embryo defective 1923 |
-0.222 |
-0.664 |
| 96 |
266336_at |
AT2G32270
|
ZIP3, zinc transporter 3 precursor |
-0.221 |
-2.253 |
| 97 |
265333_at |
AT2G18350
|
AtHB24, homeobox protein 24, HB24, homeobox protein 24, ZHD6, ZINC FINGER HOMEODOMAIN 6 |
-0.221 |
0.506 |
| 98 |
250438_at |
AT5G10580
|
unknown |
-0.219 |
-4.208 |
| 99 |
259462_at |
AT1G18940
|
[Nodulin-like / Major Facilitator Superfamily protein] |
-0.216 |
-0.798 |
| 100 |
246993_at |
AT5G67450
|
AZF1, zinc-finger protein 1, ZF1, zinc-finger protein 1 |
-0.216 |
-3.244 |
| 101 |
247487_at |
AT5G62150
|
[peptidoglycan-binding LysM domain-containing protein] |
-0.215 |
-2.903 |
| 102 |
261562_at |
AT1G01750
|
ADF11, actin depolymerizing factor 11 |
-0.214 |
-2.619 |
| 103 |
251918_at |
AT3G54040
|
[PAR1 protein] |
-0.212 |
-2.716 |
| 104 |
258760_at |
AT3G10780
|
[emp24/gp25L/p24 family/GOLD family protein] |
-0.206 |
-5.044 |
| 105 |
261937_at |
AT1G22570
|
[Major facilitator superfamily protein] |
-0.205 |
-1.707 |
| 106 |
254633_at |
AT4G18640
|
MRH1, morphogenesis of root hair 1 |
-0.204 |
-3.124 |
| 107 |
254774_at |
AT4G13440
|
[Calcium-binding EF-hand family protein] |
-0.204 |
-3.383 |
| 108 |
252879_at |
AT4G39390
|
ATNST-KT1, A. THALIANA NUCLEOTIDE SUGAR TRANSPORTER-KT 1, NST-K1, nucleotide sugar transporter-KT 1 |
-0.204 |
-1.360 |
| 109 |
264907_at |
AT2G17280
|
[Phosphoglycerate mutase family protein] |
-0.201 |
-4.536 |
| 110 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
-0.199 |
-1.848 |