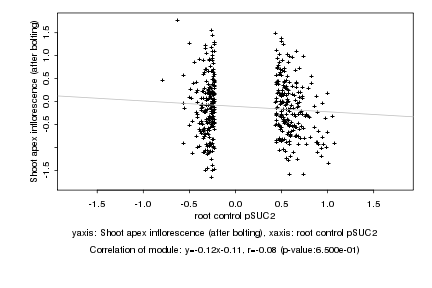
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
root control pSUC2 |
Log2 signal ratio
Shoot apex inflorescence (after bolting) |
| 1 |
248227_at |
AT5G53820
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.070 |
-0.895 |
| 2 |
247754_at |
AT5G59080
|
unknown |
1.050 |
-0.322 |
| 3 |
250031_at |
AT5G18240
|
ATMYR1, ARABIDOPSIS MYB-RELATED PROTEIN 1, MYR1, myb-related protein 1 |
1.006 |
-1.346 |
| 4 |
265025_at |
AT1G24575
|
unknown |
0.990 |
0.177 |
| 5 |
263652_at |
AT1G04330
|
unknown |
0.989 |
-0.666 |
| 6 |
260773_at |
AT1G78440
|
ATGA2OX1, Arabidopsis thaliana gibberellin 2-oxidase 1, GA2OX1, gibberellin 2-oxidase 1 |
0.985 |
-0.994 |
| 7 |
255016_at |
AT4G10120
|
ATSPS4F |
0.969 |
-0.351 |
| 8 |
261684_at |
AT1G47400
|
unknown |
0.947 |
-0.932 |
| 9 |
265769_at |
AT2G48090
|
unknown |
0.935 |
-0.773 |
| 10 |
251360_at |
AT3G61210
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.935 |
-0.022 |
| 11 |
250881_at |
AT5G04080
|
unknown |
0.926 |
-1.177 |
| 12 |
259329_at |
AT3G16360
|
AHP4, HPT phosphotransmitter 4 |
0.926 |
-1.003 |
| 13 |
267497_at |
AT2G30540
|
[Thioredoxin superfamily protein] |
0.899 |
-0.641 |
| 14 |
266597_at |
AT2G46130
|
ATWRKY43, WRKY DNA-BINDING PROTEIN 43, WRKY43, WRKY DNA-binding protein 43 |
0.898 |
-0.923 |
| 15 |
261133_at |
AT1G19715
|
[Mannose-binding lectin superfamily protein] |
0.890 |
-0.227 |
| 16 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
0.888 |
-0.882 |
| 17 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
0.887 |
-1.097 |
| 18 |
257698_at |
AT3G12730
|
[Homeodomain-like superfamily protein] |
0.878 |
0.112 |
| 19 |
258807_at |
AT3G04030
|
MYR2 |
0.869 |
-0.878 |
| 20 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
0.850 |
-0.557 |
| 21 |
246195_at |
AT4G36410
|
UBC17, ubiquitin-conjugating enzyme 17 |
0.849 |
-0.105 |
| 22 |
255626_at |
AT4G00780
|
[TRAF-like family protein] |
0.824 |
0.414 |
| 23 |
248684_at |
AT5G48485
|
DIR1, DEFECTIVE IN INDUCED RESISTANCE 1 |
0.818 |
0.557 |
| 24 |
256569_at |
AT3G19550
|
unknown |
0.810 |
-0.780 |
| 25 |
250223_at |
AT5G14070
|
ROXY2 |
0.797 |
-0.345 |
| 26 |
262921_at |
AT1G79430
|
APL, ALTERED PHLOEM DEVELOPMENT, WDY, WOODY |
0.791 |
-0.325 |
| 27 |
263370_at |
AT2G20500
|
unknown |
0.786 |
0.292 |
| 28 |
250123_at |
AT5G16530
|
PIN5, PIN-FORMED 5 |
0.770 |
-0.277 |
| 29 |
249972_at |
AT5G19040
|
ATIPT5, Arabidopsis thaliana ISOPENTENYLTRANSFERASE 5, IPT5, isopentenyltransferase 5 |
0.763 |
-0.928 |
| 30 |
245385_at |
AT4G14020
|
[Rapid alkalinization factor (RALF) family protein] |
0.761 |
-0.299 |
| 31 |
254305_at |
AT4G22200
|
AKT2, AKT2/3, potassium transport 2/3, AKT3, KT2/3, potassium transport 2/3 |
0.759 |
-0.326 |
| 32 |
262357_at |
AT1G73040
|
[Mannose-binding lectin superfamily protein] |
0.745 |
-0.789 |
| 33 |
248230_at |
AT5G53830
|
[VQ motif-containing protein] |
0.734 |
-0.570 |
| 34 |
258103_at |
AT3G23630
|
ATIPT7, ARABIDOPSIS THALIANA ISOPENTENYLTRANSFERASE 7, IPT7, isopentenyltransferase 7 |
0.733 |
-1.569 |
| 35 |
245845_at |
AT1G26150
|
AtPERK10, proline-rich extensin-like receptor kinase 10, PERK10, proline-rich extensin-like receptor kinase 10 |
0.733 |
0.983 |
| 36 |
265719_at |
AT2G03500
|
[Homeodomain-like superfamily protein] |
0.725 |
0.327 |
| 37 |
248466_at |
AT5G50720
|
ATHVA22E, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE E, HVA22E, HVA22 homologue E |
0.723 |
0.082 |
| 38 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
0.718 |
0.124 |
| 39 |
246429_at |
AT5G17450
|
HIPP21, heavy metal associated isoprenylated plant protein 21 |
0.717 |
-0.268 |
| 40 |
246440_at |
AT5G17650
|
[glycine/proline-rich protein] |
0.714 |
-0.214 |
| 41 |
250650_at |
AT5G06850
|
FTIP1, FT-interacting protein 1 |
0.711 |
-0.530 |
| 42 |
249094_at |
AT5G43890
|
SUPER1, SUPPRESSOR OF ER 1, YUC5, YUCCA5 |
0.701 |
-0.788 |
| 43 |
253002_at |
AT4G38530
|
ATPLC1, phospholipase C1, PLC1, phospholipase C1 |
0.700 |
-0.238 |
| 44 |
261574_at |
AT1G01190
|
CYP78A8, cytochrome P450, family 78, subfamily A, polypeptide 8 |
0.700 |
0.204 |
| 45 |
266325_at |
AT2G46630
|
unknown |
0.693 |
0.724 |
| 46 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
0.693 |
-0.009 |
| 47 |
248491_at |
AT5G51010
|
[Rubredoxin-like superfamily protein] |
0.686 |
0.105 |
| 48 |
260076_at |
AT1G73630
|
[EF hand calcium-binding protein family] |
0.686 |
-0.597 |
| 49 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
0.684 |
-0.745 |
| 50 |
258170_at |
AT3G21600
|
[Senescence/dehydration-associated protein-related] |
0.682 |
-0.918 |
| 51 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
0.682 |
0.207 |
| 52 |
260672_at |
AT1G19480
|
[DNA glycosylase superfamily protein] |
0.680 |
-0.476 |
| 53 |
247282_at |
AT5G64240
|
AtMC3, metacaspase 3, AtMCP1a, metacaspase 1a, MC3, metacaspase 3, MCP1a, metacaspase 1a |
0.676 |
-0.108 |
| 54 |
265939_at |
AT2G19650
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.674 |
-0.272 |
| 55 |
261545_at |
AT1G63530
|
unknown |
0.670 |
-1.260 |
| 56 |
245384_at |
AT4G16790
|
[hydroxyproline-rich glycoprotein family protein] |
0.667 |
-0.119 |
| 57 |
248443_at |
AT5G51310
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.664 |
-0.037 |
| 58 |
250869_at |
AT5G03840
|
TFL-1, TERMINAL FLOWER 1, TFL1, TERMINAL FLOWER 1 |
0.660 |
0.406 |
| 59 |
247964_at |
AT5G56600
|
PFN3, PROFILIN 3, PRF3, profilin 3 |
0.660 |
0.325 |
| 60 |
261100_at |
AT1G63020
|
NRPD1, NRPD1A, nuclear RNA polymerase D1A, POL IVA, NUCLEAR RNA POLYMERASE D 1A, SDE4, SMD2, SILENCING MOVEMENT DEFICIENT 2 |
0.658 |
1.092 |
| 61 |
254362_at |
AT4G22160
|
unknown |
0.658 |
0.428 |
| 62 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.655 |
-0.565 |
| 63 |
245990_at |
AT5G20640
|
unknown |
0.653 |
-0.721 |
| 64 |
254374_at |
AT4G21780
|
unknown |
0.653 |
-0.757 |
| 65 |
257466_at |
AT1G62840
|
unknown |
0.650 |
-0.756 |
| 66 |
267120_at |
AT2G23530
|
[Zinc-finger domain of monoamine-oxidase A repressor R1] |
0.649 |
0.034 |
| 67 |
245692_at |
AT5G04150
|
BHLH101 |
0.647 |
-0.556 |
| 68 |
248476_at |
AT5G50890
|
[alpha/beta-Hydrolases superfamily protein] |
0.646 |
0.680 |
| 69 |
258196_at |
AT3G13980
|
unknown |
0.641 |
-0.312 |
| 70 |
249401_at |
AT5G40260
|
AtSWEET8, RPG1, RUPTURED POLLEN GRAIN1, SWEET8 |
0.636 |
-0.504 |
| 71 |
245758_at |
AT1G32240
|
KAN2, KANADI 2 |
0.635 |
0.133 |
| 72 |
257398_at |
AT2G01990
|
unknown |
0.631 |
-0.280 |
| 73 |
251154_at |
AT3G63110
|
ATIPT3, isopentenyltransferase 3, IPT3, isopentenyltransferase 3 |
0.630 |
-0.150 |
| 74 |
260475_at |
AT1G11080
|
scpl31, serine carboxypeptidase-like 31 |
0.629 |
-1.107 |
| 75 |
262940_at |
AT1G79520
|
[Cation efflux family protein] |
0.625 |
-0.623 |
| 76 |
259694_at |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
0.618 |
-0.524 |
| 77 |
247959_at |
AT5G57080
|
unknown |
0.617 |
0.158 |
| 78 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
0.616 |
-0.263 |
| 79 |
247643_at |
AT5G60450
|
ARF4, auxin response factor 4 |
0.614 |
0.969 |
| 80 |
265926_at |
AT2G18600
|
[Ubiquitin-conjugating enzyme family protein] |
0.611 |
0.329 |
| 81 |
261596_at |
AT1G33080
|
[MATE efflux family protein] |
0.602 |
-0.069 |
| 82 |
250258_at |
AT5G13790
|
AGL15, AGAMOUS-like 15 |
0.601 |
-0.635 |
| 83 |
250803_at |
AT5G04980
|
[DNAse I-like superfamily protein] |
0.600 |
-0.451 |
| 84 |
256319_at |
AT1G35910
|
TPPD, trehalose-6-phosphate phosphatase D |
0.597 |
-1.177 |
| 85 |
252070_at |
AT3G51680
|
AtSDR2, SDR2, short-chain dehydrogenase/reductase 2 |
0.595 |
-0.736 |
| 86 |
261212_at |
AT1G12880
|
atnudt12, nudix hydrolase homolog 12, NUDT12, nudix hydrolase homolog 12 |
0.594 |
-0.149 |
| 87 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
0.589 |
-0.708 |
| 88 |
261504_at |
AT1G71692
|
AGL12, AGAMOUS-like 12, XAL1, XAANTAL1 |
0.588 |
-0.756 |
| 89 |
254551_at |
AT4G19840
|
ATPP2-A1, phloem protein 2-A1, ATPP2A-1, phloem protein 2-A1, PP2-A1, phloem protein 2-A1 |
0.587 |
-0.501 |
| 90 |
248590_at |
AT5G49660
|
XIP1, XYLEM INTERMIXED WITH PHLOEM 1 |
0.586 |
0.419 |
| 91 |
259832_at |
AT1G69580
|
[Homeodomain-like superfamily protein] |
0.584 |
-0.216 |
| 92 |
256537_at |
AT1G33340
|
[ENTH/ANTH/VHS superfamily protein] |
0.583 |
-0.930 |
| 93 |
248484_at |
AT5G51030
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.582 |
-0.863 |
| 94 |
260334_at |
AT1G70510
|
ATK1, ARABIDOPSIS THALIANA KN 1, KNAT2, KNOTTED-like from Arabidopsis thaliana 2 |
0.579 |
0.997 |
| 95 |
252163_at |
AT3G50610
|
unknown |
0.578 |
-0.544 |
| 96 |
264281_at |
AT1G61830
|
[pseudogene] |
0.577 |
-1.587 |
| 97 |
248208_at |
AT5G53980
|
ATHB52, homeobox protein 52, HB52, homeobox protein 52 |
0.575 |
-0.409 |
| 98 |
251207_at |
AT3G63050
|
unknown |
0.573 |
-0.498 |
| 99 |
263297_at |
AT2G15310
|
ARFB1A, ADP-ribosylation factor B1A, ATARFB1A, ADP-ribosylation factor B1A |
0.573 |
-0.662 |
| 100 |
251289_at |
AT3G61830
|
ARF18, auxin response factor 18 |
0.571 |
0.021 |
| 101 |
263136_at |
AT1G78580
|
ATTPS1, trehalose-6-phosphate synthase, TPS1, trehalose-6-phosphate synthase, TPS1, TREHALOSE-6-PHOSPHATE SYNTHASE 1 |
0.570 |
0.248 |
| 102 |
263258_at |
AT1G10540
|
ATNAT8, NAT8, nucleobase-ascorbate transporter 8 |
0.569 |
-0.161 |
| 103 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
0.568 |
-1.273 |
| 104 |
246653_at |
AT5G35200
|
[ENTH/ANTH/VHS superfamily protein] |
0.566 |
0.852 |
| 105 |
248172_at |
AT5G54660
|
[HSP20-like chaperones superfamily protein] |
0.565 |
0.190 |
| 106 |
259838_at |
AT1G52220
|
CURT1C, CURVATURE THYLAKOID 1C |
0.563 |
0.560 |
| 107 |
265204_at |
AT2G36650
|
unknown |
0.561 |
-0.010 |
| 108 |
262905_at |
AT1G59730
|
ATH7, thioredoxin H-type 7, TH7, thioredoxin H-type 7 |
0.561 |
-0.496 |
| 109 |
252501_at |
AT3G46880
|
unknown |
0.560 |
-0.372 |
| 110 |
247340_at |
AT5G63700
|
[zinc ion binding] |
0.560 |
-0.008 |
| 111 |
253243_at |
AT4G34560
|
unknown |
0.558 |
0.050 |
| 112 |
266321_at |
AT2G46660
|
CYP78A6, cytochrome P450, family 78, subfamily A, polypeptide 6, EOD3, enhancer of da1-1 |
0.557 |
0.277 |
| 113 |
266458_at |
AT2G47710
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.556 |
1.041 |
| 114 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
0.556 |
-0.704 |
| 115 |
259058_at |
AT3G03470
|
CYP89A9, cytochrome P450, family 87, subfamily A, polypeptide 9 |
0.553 |
0.138 |
| 116 |
250186_at |
AT5G14500
|
[aldose 1-epimerase family protein] |
0.552 |
-0.156 |
| 117 |
246279_at |
AT4G36740
|
ATHB40, homeobox protein 40, HB-5, HB40, homeobox protein 40 |
0.552 |
-1.233 |
| 118 |
247775_at |
AT5G58690
|
ATPLC5, PLC5, phosphatidylinositol-speciwc phospholipase C5 |
0.551 |
-0.262 |
| 119 |
259864_at |
AT1G72800
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.548 |
-0.017 |
| 120 |
261033_at |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
0.548 |
-0.045 |
| 121 |
264532_at |
AT1G55740
|
AtSIP1, seed imbibition 1, RS1, raffinose synthase 1, SIP1, seed imbibition 1 |
0.547 |
0.339 |
| 122 |
252740_at |
AT3G43270
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.546 |
0.471 |
| 123 |
259765_at |
AT1G64370
|
unknown |
0.544 |
-0.883 |
| 124 |
247357_at |
AT5G63710
|
[Leucine-rich repeat protein kinase family protein] |
0.542 |
-0.415 |
| 125 |
247902_at |
AT5G57350
|
AHA3, H(+)-ATPase 3, ATAHA3, ARABIDOPSIS THALIANA ARABIDOPSIS H(+)-ATPASE, HA3, H(+)-ATPase 3 |
0.542 |
0.300 |
| 126 |
258287_at |
AT3G15990
|
SULTR3;4, sulfate transporter 3;4 |
0.542 |
-0.452 |
| 127 |
247035_at |
AT5G67110
|
ALC, ALCATRAZ |
0.537 |
-0.065 |
| 128 |
257375_at |
AT2G38640
|
unknown |
0.533 |
-1.086 |
| 129 |
254998_at |
AT4G09760
|
[Protein kinase superfamily protein] |
0.532 |
0.178 |
| 130 |
252700_at |
AT3G43690
|
[copia-like retrotransposon family protein, has a 1.4e-29 P-value blast match to gb|AAG52950.1| putative envelope protein (Endovir1-1) (Arabidopsis thaliana) (Ty1_Copia-family)] |
0.530 |
-0.473 |
| 131 |
265825_at |
AT2G35635
|
RUB2, RELATED TO UBIQUITIN 2, UBQ7, ubiquitin 7 |
0.529 |
0.423 |
| 132 |
250107_at |
AT5G15330
|
ATSPX4, ARABIDOPSIS THALIANA SPX DOMAIN GENE 4, SPX4, SPX domain gene 4 |
0.529 |
0.448 |
| 133 |
251243_at |
AT3G61870
|
unknown |
0.527 |
0.730 |
| 134 |
265395_at |
AT2G20850
|
SRF1, STRUBBELIG-receptor family 1 |
0.524 |
0.658 |
| 135 |
245650_at |
AT1G24735
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.524 |
0.314 |
| 136 |
247940_at |
AT5G57190
|
PSD2, phosphatidylserine decarboxylase 2 |
0.523 |
-0.827 |
| 137 |
265357_at |
AT2G16740
|
UBC29, ubiquitin-conjugating enzyme 29 |
0.522 |
-0.411 |
| 138 |
254559_at |
AT4G19200
|
[proline-rich family protein] |
0.521 |
-0.346 |
| 139 |
261467_at |
AT1G28520
|
ATVOZ1, VASCULAR PLANT ONE ZINC FINGER PROTEIN, VOZ1, vascular plant one zinc finger protein |
0.520 |
-0.161 |
| 140 |
251768_at |
AT3G55940
|
[Phosphoinositide-specific phospholipase C family protein] |
0.519 |
0.139 |
| 141 |
247656_at |
AT5G59890
|
ADF4, actin depolymerizing factor 4, ATADF4 |
0.519 |
-0.011 |
| 142 |
257232_at |
AT3G16500
|
IAA26, indole-3-acetic acid inducible 26, PAP1, phytochrome-associated protein 1 |
0.519 |
1.245 |
| 143 |
264693_at |
AT1G69970
|
CLE26, CLAVATA3/ESR-RELATED 26 |
0.517 |
-1.038 |
| 144 |
260805_at |
AT1G78320
|
ATGSTU23, glutathione S-transferase TAU 23, GSTU23, glutathione S-transferase TAU 23 |
0.517 |
-0.374 |
| 145 |
264204_at |
AT1G22710
|
ATSUC2, ARABIDOPSIS THALIANA SUCROSE-PROTON SYMPORTER 2, SUC2, sucrose-proton symporter 2, SUT1, SUCROSE TRANSPORTER 1 |
0.517 |
0.361 |
| 146 |
255563_at |
AT4G01740
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.517 |
-0.417 |
| 147 |
246215_at |
AT4G37180
|
[Homeodomain-like superfamily protein] |
0.509 |
0.689 |
| 148 |
265546_at |
AT2G28240
|
[ATP-dependent helicase family protein] |
0.507 |
0.009 |
| 149 |
250471_at |
AT5G10170
|
ATMIPS3, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 3, MIPS3, myo-inositol-1-phosphate synthase 3 |
0.506 |
0.258 |
| 150 |
248967_at |
AT5G45350
|
[proline-rich family protein] |
0.506 |
0.179 |
| 151 |
259857_at |
AT1G68430
|
unknown |
0.504 |
-0.632 |
| 152 |
251301_at |
AT3G61880
|
CYP78A9, cytochrome p450 78a9 |
0.503 |
-0.820 |
| 153 |
258879_at |
AT3G03270
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.502 |
0.906 |
| 154 |
246724_at |
AT5G29000
|
PHL1, PHR1-like 1 |
0.502 |
-0.849 |
| 155 |
264145_at |
AT1G79310
|
AtMC7, metacaspase 7, AtMCP2a, metacaspase 2a, MC7, metacaspase 7, MCP2a, metacaspase 2a |
0.502 |
0.149 |
| 156 |
262162_at |
AT1G78020
|
unknown |
0.499 |
1.389 |
| 157 |
261610_at |
AT1G49560
|
[Homeodomain-like superfamily protein] |
0.497 |
-0.214 |
| 158 |
247582_at |
AT5G60760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.494 |
-0.297 |
| 159 |
253393_at |
AT4G32690
|
ATGLB3, ARABIDOPSIS HEMOGLOBIN 3, GLB3, hemoglobin 3 |
0.494 |
-0.556 |
| 160 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
0.494 |
-0.026 |
| 161 |
246833_at |
AT5G26620
|
unknown |
0.494 |
-0.852 |
| 162 |
264837_at |
AT1G03600
|
PSB27 |
0.493 |
1.322 |
| 163 |
258456_at |
AT3G22420
|
ATWNK2, ARABIDOPSIS THALIANA WITH NO K 2, WNK2, with no lysine (K) kinase 2, ZIK3 |
0.492 |
0.154 |
| 164 |
265494_at |
AT2G15680
|
AtCML30, CML30, calmodulin-like 30 |
0.492 |
-0.333 |
| 165 |
245728_at |
AT1G73340
|
[Cytochrome P450 superfamily protein] |
0.491 |
-0.264 |
| 166 |
255958_at |
AT1G22150
|
SULTR1;3, sulfate transporter 1;3 |
0.490 |
-0.842 |
| 167 |
257054_at |
AT3G15353
|
ATMT3, MT3, metallothionein 3 |
0.490 |
0.583 |
| 168 |
245701_at |
AT5G04140
|
FD-GOGAT, FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE, GLS1, FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE 1, GLU1, glutamate synthase 1, GLUS |
0.489 |
0.112 |
| 169 |
256093_at |
AT1G20823
|
[RING/U-box superfamily protein] |
0.487 |
-0.305 |
| 170 |
260007_at |
AT1G67870
|
[glycine-rich protein] |
0.486 |
0.649 |
| 171 |
245499_at |
AT4G16480
|
ATINT4, INT4, inositol transporter 4 |
0.485 |
0.007 |
| 172 |
245318_at |
AT4G16980
|
[arabinogalactan-protein family] |
0.483 |
0.703 |
| 173 |
253779_at |
AT4G28490
|
HAE, HAESA, RLK5, RECEPTOR-LIKE PROTEIN KINASE 5 |
0.483 |
-0.325 |
| 174 |
249126_at |
AT5G43380
|
TOPP6, type one serine/threonine protein phosphatase 6 |
0.482 |
0.153 |
| 175 |
245186_at |
AT1G67710
|
ARR11, response regulator 11 |
0.479 |
-0.605 |
| 176 |
255572_at |
AT4G01050
|
TROL, thylakoid rhodanese-like |
0.479 |
-0.057 |
| 177 |
259423_at |
AT1G13880
|
[ELM2 domain-containing protein] |
0.479 |
0.649 |
| 178 |
264512_at |
AT1G09575
|
unknown |
0.478 |
-0.536 |
| 179 |
266369_at |
AT2G41370
|
BOP2, BLADE ON PETIOLE2 |
0.478 |
1.043 |
| 180 |
260315_at |
AT1G63820
|
[CCT motif family protein] |
0.477 |
-0.798 |
| 181 |
252716_at |
AT3G43920
|
ATDCL3, DICER-LIKE 3, DCL3, dicer-like 3 |
0.477 |
0.871 |
| 182 |
245998_at |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
0.475 |
0.189 |
| 183 |
257715_at |
AT3G12750
|
AtZIP1, ZIP1, zinc transporter 1 precursor |
0.474 |
0.603 |
| 184 |
248969_at |
AT5G45310
|
unknown |
0.474 |
-0.848 |
| 185 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
0.473 |
-0.506 |
| 186 |
262778_at |
AT1G13050
|
unknown |
0.472 |
-1.052 |
| 187 |
251062_at |
AT5G01840
|
ATOFP1, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 1, OFP1, ovate family protein 1 |
0.470 |
-0.816 |
| 188 |
262967_at |
AT1G75730
|
unknown |
0.465 |
0.392 |
| 189 |
257476_at |
AT1G80960
|
[F-box and Leucine Rich Repeat domains containing protein] |
0.464 |
0.452 |
| 190 |
251491_at |
AT3G59480
|
[pfkB-like carbohydrate kinase family protein] |
0.463 |
-0.492 |
| 191 |
254188_at |
AT4G23920
|
ATUGE2, UDP-GLC 4-EPIMERASE 2, UGE2, UDP-D-glucose/UDP-D-galactose 4-epimerase 2 |
0.462 |
0.207 |
| 192 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
0.462 |
0.258 |
| 193 |
257600_at |
AT3G24770
|
CLE41, CLAVATA3/ESR-RELATED 41 |
0.462 |
0.790 |
| 194 |
263314_at |
AT2G05760
|
[Xanthine/uracil permease family protein] |
0.456 |
0.725 |
| 195 |
248550_at |
AT5G50210
|
OLD5, ONSET OF LEAF DEATH 5, QS, quinolinate synthase, SUFE3, SULFUR E 3 |
0.451 |
0.411 |
| 196 |
247304_at |
AT5G63850
|
AAP4, amino acid permease 4 |
0.451 |
-0.123 |
| 197 |
257916_at |
AT3G23210
|
bHLH34, basic Helix-Loop-Helix 34 |
0.449 |
0.750 |
| 198 |
256681_at |
AT3G52340
|
ATSPP2, SUCROSE-PHOSPHATASE 2, SPP2, sucrose-6F-phosphate phosphohydrolase 2 |
0.448 |
0.453 |
| 199 |
262728_at |
AT1G75820
|
ATCLV1, CLV1, CLAVATA 1, FAS3, FASCIATA 3, FLO5, FLOWER DEVELOPMENT 5 |
0.448 |
0.188 |
| 200 |
246622_at |
AT5G36250
|
PP2C74, protein phosphatase 2C 74 |
0.447 |
0.951 |
| 201 |
264836_at |
AT1G03610
|
unknown |
0.445 |
0.573 |
| 202 |
251058_at |
AT5G01790
|
unknown |
0.445 |
-0.404 |
| 203 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
0.445 |
-0.151 |
| 204 |
259953_at |
AT1G74810
|
BOR5, REQUIRES HIGH BORON 5 |
0.443 |
-0.301 |
| 205 |
255645_at |
AT4G00880
|
SAUR31, SMALL AUXIN UPREGULATED RNA 31 |
0.443 |
0.383 |
| 206 |
247318_at |
AT5G63990
|
[Inositol monophosphatase family protein] |
0.443 |
0.420 |
| 207 |
261518_at |
AT1G71695
|
[Peroxidase superfamily protein] |
0.441 |
1.120 |
| 208 |
252786_at |
AT3G42670
|
CHR38, chromatin remodeling 38, CLSY, CLASSY1, CLSY1, CLASSY 1 |
0.438 |
0.007 |
| 209 |
251006_at |
AT5G02600
|
NAKR1, SODIUM POTASSIUM ROOT DEFECTIVE 1, NPCC6, nuclear-enriched phloem companion cell gene 6 |
0.437 |
-0.255 |
| 210 |
245202_at |
AT1G67720
|
[Leucine-rich repeat protein kinase family protein] |
0.437 |
-0.481 |
| 211 |
251592_at |
AT3G57670
|
NTT, NO TRANSMITTING TRACT, WIP2, WIP domain protein 2 |
0.437 |
-0.832 |
| 212 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.436 |
0.083 |
| 213 |
253791_at |
AT4G28640
|
IAA11, indole-3-acetic acid inducible 11 |
0.435 |
0.585 |
| 214 |
264340_at |
AT1G70280
|
[NHL domain-containing protein] |
0.435 |
-0.264 |
| 215 |
265186_at |
AT1G23560
|
unknown |
0.435 |
-0.882 |
| 216 |
259426_at |
AT1G01470
|
LEA14, LATE EMBRYOGENESIS ABUNDANT 14, LSR3, LIGHT STRESS-REGULATED 3 |
0.434 |
-0.505 |
| 217 |
259182_at |
AT3G01750
|
[Ankyrin repeat family protein] |
0.433 |
-0.011 |
| 218 |
260307_at |
AT1G70620
|
[cyclin-related] |
0.432 |
1.504 |
| 219 |
244960_at |
ATCG01020
|
RPL32, ribosomal protein L32 |
-0.796 |
0.467 |
| 220 |
258675_at |
AT3G08770
|
LTP6, lipid transfer protein 6 |
-0.639 |
1.787 |
| 221 |
265057_at |
AT1G52140
|
unknown |
-0.569 |
0.581 |
| 222 |
244980_at |
ATCG00760
|
RPL36, ribosomal protein L36 |
-0.568 |
-0.028 |
| 223 |
264403_at |
AT2G25150
|
[HXXXD-type acyl-transferase family protein] |
-0.565 |
-0.913 |
| 224 |
244969_at |
ATCG00650
|
RPS18, ribosomal protein S18 |
-0.553 |
-0.132 |
| 225 |
261221_at |
AT1G19960
|
unknown |
-0.509 |
1.276 |
| 226 |
247337_at |
AT5G63660
|
LCR74, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 74, PDF2.5 |
-0.509 |
0.094 |
| 227 |
267343_at |
AT2G44260
|
unknown |
-0.507 |
-0.522 |
| 228 |
245017_at |
ATCG00510
|
PSAI, photsystem I subunit I |
-0.491 |
0.266 |
| 229 |
264029_at |
AT2G03720
|
MRH6, morphogenesis of root hair 6 |
-0.486 |
0.087 |
| 230 |
251481_at |
AT3G59730
|
LecRK-V.6, L-type lectin receptor kinase V.6 |
-0.476 |
-1.122 |
| 231 |
247657_at |
AT5G59845
|
[Gibberellin-regulated family protein] |
-0.470 |
-0.419 |
| 232 |
254041_at |
AT4G25830
|
[Uncharacterised protein family (UPF0497)] |
-0.457 |
0.409 |
| 233 |
245571_at |
AT4G14695
|
[Uncharacterised protein family (UPF0041)] |
-0.455 |
0.858 |
| 234 |
244985_at |
ATCG00810
|
RPL22, ribosomal protein L22 |
-0.433 |
0.423 |
| 235 |
257334_at |
ATMG01370
|
ORF111D |
-0.428 |
0.203 |
| 236 |
258765_at |
AT3G10710
|
RHS12, root hair specific 12 |
-0.424 |
-0.744 |
| 237 |
265924_at |
AT2G18620
|
[Terpenoid synthases superfamily protein] |
-0.424 |
-0.230 |
| 238 |
259609_at |
AT1G52410
|
AtTSA1, TSA1, TSK-associating protein 1 |
-0.421 |
-0.018 |
| 239 |
259199_at |
AT3G08980
|
[Peptidase S24/S26A/S26B/S26C family protein] |
-0.418 |
-0.346 |
| 240 |
251019_at |
AT5G02420
|
unknown |
-0.417 |
-0.262 |
| 241 |
249037_at |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
-0.416 |
0.242 |
| 242 |
258178_at |
AT3G21680
|
unknown |
-0.414 |
-0.997 |
| 243 |
254494_at |
AT4G20050
|
QRT3, QUARTET 3 |
-0.399 |
-0.475 |
| 244 |
267217_at |
AT2G02610
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.399 |
-0.962 |
| 245 |
256442_at |
AT3G10930
|
unknown |
-0.395 |
-0.602 |
| 246 |
260547_at |
AT2G43550
|
[Scorpion toxin-like knottin superfamily protein] |
-0.394 |
0.936 |
| 247 |
251535_at |
AT3G58540
|
unknown |
-0.387 |
-0.645 |
| 248 |
254644_at |
AT4G18510
|
CLE2, CLAVATA3/ESR-related 2 |
-0.377 |
-0.085 |
| 249 |
266276_at |
AT2G29330
|
TRI, tropinone reductase |
-0.375 |
0.077 |
| 250 |
257670_at |
AT3G20340
|
unknown |
-0.373 |
-0.420 |
| 251 |
266649_at |
AT2G25810
|
TIP4;1, tonoplast intrinsic protein 4;1 |
-0.373 |
-0.137 |
| 252 |
250157_at |
AT5G15180
|
[Peroxidase superfamily protein] |
-0.372 |
-0.179 |
| 253 |
254510_at |
AT4G20210
|
TPS08, terpene synthase 8 |
-0.371 |
-0.462 |
| 254 |
249061_at |
AT5G44550
|
[Uncharacterised protein family (UPF0497)] |
-0.369 |
-0.685 |
| 255 |
246920_at |
AT5G25090
|
AtENODL13, ENODL13, early nodulin-like protein 13 |
-0.368 |
0.239 |
| 256 |
259274_at |
AT3G01220
|
ATHB20, homeobox protein 20, HB20, homeobox protein 20 |
-0.366 |
-0.485 |
| 257 |
250882_at |
AT5G04000
|
unknown |
-0.364 |
-0.053 |
| 258 |
252172_at |
AT3G50640
|
unknown |
-0.363 |
-0.483 |
| 259 |
255591_at |
AT4G01630
|
ATEXP17, EXPANSIN 17, ATEXPA17, expansin A17, ATHEXP ALPHA 1.13, EXPA17, expansin A17 |
-0.360 |
-0.127 |
| 260 |
261958_at |
AT1G64500
|
[Glutaredoxin family protein] |
-0.354 |
-0.597 |
| 261 |
266737_at |
AT2G47140
|
AtSDR5, SDR5, short-chain dehydrogenase reductase 5 |
-0.353 |
-0.650 |
| 262 |
263689_at |
AT1G26820
|
RNS3, ribonuclease 3 |
-0.352 |
0.078 |
| 263 |
245001_at |
ATCG00220
|
PSBM, photosystem II reaction center protein M |
-0.352 |
0.592 |
| 264 |
248912_at |
AT5G45670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.351 |
0.904 |
| 265 |
249108_at |
AT5G43690
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.351 |
-0.628 |
| 266 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
-0.348 |
0.220 |
| 267 |
262643_at |
AT1G62770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.347 |
-0.805 |
| 268 |
251293_at |
AT3G61930
|
unknown |
-0.347 |
-1.108 |
| 269 |
263387_at |
AT2G40160
|
TBL30 |
-0.345 |
-0.756 |
| 270 |
245009_at |
ATCG00380
|
RPS4, chloroplast ribosomal protein S4 |
-0.345 |
0.641 |
| 271 |
255111_at |
AT4G08780
|
[Peroxidase superfamily protein] |
-0.343 |
-0.569 |
| 272 |
259735_at |
AT1G64405
|
unknown |
-0.342 |
-0.716 |
| 273 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
-0.341 |
-1.109 |
| 274 |
250746_at |
AT5G05880
|
[UDP-Glycosyltransferase superfamily protein] |
-0.340 |
-0.760 |
| 275 |
266334_at |
AT2G32380
|
[Transmembrane protein 97, predicted] |
-0.339 |
-0.212 |
| 276 |
264581_at |
AT1G05210
|
[Transmembrane protein 97, predicted] |
-0.339 |
0.284 |
| 277 |
244906_at |
ATMG00690
|
ORF240A |
-0.336 |
-0.230 |
| 278 |
266307_at |
AT2G27000
|
CYP705A8, cytochrome P450, family 705, subfamily A, polypeptide 8 |
-0.335 |
-1.486 |
| 279 |
260005_at |
AT1G67920
|
unknown |
-0.335 |
-0.192 |
| 280 |
248763_at |
AT5G47550
|
[Cystatin/monellin superfamily protein] |
-0.335 |
0.704 |
| 281 |
253372_at |
AT4G33220
|
ATPME44, A. THALIANA PECTIN METHYLESTERASE 44, PME44, pectin methylesterase 44 |
-0.334 |
1.234 |
| 282 |
246548_at |
AT5G14910
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.334 |
0.182 |
| 283 |
266415_at |
AT2G38530
|
cdf3, cell growth defect factor-3, LP2, LTP2, lipid transfer protein 2 |
-0.333 |
1.177 |
| 284 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
-0.331 |
-1.049 |
| 285 |
254391_at |
AT4G21590
|
ENDO3, endonuclease 3 |
-0.329 |
-0.222 |
| 286 |
263127_at |
AT1G78610
|
MSL6, mechanosensitive channel of small conductance-like 6 |
-0.329 |
-0.203 |
| 287 |
266974_at |
AT2G39370
|
MAKR4, MEMBRANE-ASSOCIATED KINASE REGULATOR 4 |
-0.326 |
-0.711 |
| 288 |
244967_at |
ATCG00630
|
PSAJ |
-0.324 |
0.717 |
| 289 |
254258_at |
AT4G23410
|
TET5, tetraspanin5 |
-0.324 |
-0.433 |
| 290 |
245889_at |
AT5G09480
|
[hydroxyproline-rich glycoprotein family protein] |
-0.323 |
-0.926 |
| 291 |
250200_at |
AT5G14130
|
[Peroxidase superfamily protein] |
-0.321 |
-0.075 |
| 292 |
262875_at |
AT1G64970
|
G-TMT, gamma-tocopherol methyltransferase, TMT1, VTE4, VITAMIN E DEFICIENT 4 |
-0.318 |
0.447 |
| 293 |
260332_at |
AT1G70470
|
unknown |
-0.317 |
-0.077 |
| 294 |
261999_at |
AT1G33800
|
AtGXMT1, GXM1, glucuronoxylan methyltransferase 1, GXMT1, glucuronoxylan methyltransferase 1 |
-0.316 |
-0.391 |
| 295 |
246765_at |
AT5G27330
|
[Prefoldin chaperone subunit family protein] |
-0.315 |
1.067 |
| 296 |
255608_at |
AT4G01140
|
unknown |
-0.315 |
0.083 |
| 297 |
245304_at |
AT4G15630
|
[Uncharacterised protein family (UPF0497)] |
-0.315 |
0.042 |
| 298 |
253725_at |
AT4G29340
|
PRF4, profilin 4 |
-0.314 |
-1.062 |
| 299 |
263376_at |
AT2G20520
|
FLA6, FASCICLIN-like arabinogalactan 6 |
-0.312 |
-1.143 |
| 300 |
265050_at |
AT1G52070
|
[Mannose-binding lectin superfamily protein] |
-0.312 |
-0.688 |
| 301 |
264696_at |
AT1G70230
|
AXY4, ALTERED XYLOGLUCAN 4, TBL27, TRICHOME BIREFRINGENCE-LIKE 27 |
-0.311 |
-0.613 |
| 302 |
263616_at |
AT2G04680
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.310 |
-1.445 |
| 303 |
250455_at |
AT5G09980
|
PROPEP4, elicitor peptide 4 precursor |
-0.307 |
-0.165 |
| 304 |
255732_at |
AT1G25450
|
CER60, ECERIFERUM 60, KCS5, 3-ketoacyl-CoA synthase 5 |
-0.305 |
-0.472 |
| 305 |
254110_at |
AT4G25260
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.304 |
-0.973 |
| 306 |
244978_at |
ATCG00740
|
RPOA, RNA polymerase subunit alpha |
-0.301 |
0.655 |
| 307 |
252458_at |
AT3G47210
|
unknown |
-0.301 |
-0.709 |
| 308 |
256926_at |
AT3G22540
|
unknown |
-0.299 |
0.279 |
| 309 |
252232_at |
AT3G49760
|
AtbZIP5, basic leucine-zipper 5, bZIP5, basic leucine-zipper 5 |
-0.299 |
0.193 |
| 310 |
256157_at |
AT1G13580
|
LAG13, LAG1 longevity assurance homolog 3, LOH3, LAG One Homologue 3 |
-0.299 |
0.244 |
| 311 |
250224_at |
AT5G14150
|
unknown |
-0.297 |
0.199 |
| 312 |
245172_at |
AT2G47540
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.296 |
-0.065 |
| 313 |
259325_at |
AT3G05320
|
[O-fucosyltransferase family protein] |
-0.296 |
-0.584 |
| 314 |
264288_at |
AT1G62045
|
unknown |
-0.295 |
0.389 |
| 315 |
258617_at |
AT3G03000
|
[EF hand calcium-binding protein family] |
-0.295 |
-1.123 |
| 316 |
257113_at |
AT3G20130
|
CYP705A22, cytochrome P450, family 705, subfamily A, polypeptide 22, GPS1, gravity persistence signal 1 |
-0.294 |
-0.361 |
| 317 |
264124_at |
AT1G79360
|
ATOCT2, organic cation/carnitine transporter 2, OCT2, ORGANIC CATION TRANSPORTER 2, OCT2, organic cation/carnitine transporter 2 |
-0.293 |
0.047 |
| 318 |
255005_at |
AT4G09990
|
GXM2, glucuronoxylan methyltransferase 2 |
-0.292 |
-0.962 |
| 319 |
245612_at |
AT4G14440
|
ATECI3, ARABIDOPSIS THALIANA DELTA(3), DELTA(2)-ENOYL COA ISOMERASE 3, ECI3, DELTA(3), DELTA(2)-ENOYL COA ISOMERASE 3, HCD1, 3-hydroxyacyl-CoA dehydratase 1 |
-0.290 |
0.904 |
| 320 |
267618_at |
AT2G26760
|
CYCB1;4, Cyclin B1;4 |
-0.289 |
0.928 |
| 321 |
267191_at |
AT2G44110
|
ATMLO15, MILDEW RESISTANCE LOCUS O 15, MLO15, MILDEW RESISTANCE LOCUS O 15 |
-0.289 |
-0.764 |
| 322 |
266356_at |
AT2G32300
|
UCC1, uclacyanin 1 |
-0.289 |
-1.097 |
| 323 |
266708_at |
AT2G03200
|
[Eukaryotic aspartyl protease family protein] |
-0.288 |
-0.946 |
| 324 |
264527_at |
AT1G30760
|
[FAD-binding Berberine family protein] |
-0.288 |
-0.307 |
| 325 |
262730_at |
AT1G16390
|
ATOCT3, organic cation/carnitine transporter 3, OCT3, organic cation/carnitine transporter 3 |
-0.288 |
-0.398 |
| 326 |
244968_at |
ATCG00640
|
RPL33, ribosomal protein L33 |
-0.287 |
0.316 |
| 327 |
245004_at |
ATCG00300
|
YCF9 |
-0.287 |
0.170 |
| 328 |
259287_at |
AT3G11490
|
[rac GTPase activating protein] |
-0.286 |
0.158 |
| 329 |
261384_at |
AT1G05440
|
[C-8 sterol isomerases] |
-0.286 |
0.233 |
| 330 |
259018_at |
AT3G07390
|
AIR12, Auxin-Induced in Root cultures 12 |
-0.286 |
0.230 |
| 331 |
263005_at |
AT1G54540
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.285 |
-0.907 |
| 332 |
254629_at |
AT4G18425
|
unknown |
-0.284 |
-0.457 |
| 333 |
254608_at |
AT4G18910
|
ATNLM2, NOD26-LIKE INTRINSIC PROTEIN 2, NIP1;2, NOD26-like intrinsic protein 1;2, NLM2, NOD26-LIKE INTRINSIC PROTEIN 2 |
-0.283 |
0.085 |
| 334 |
248790_at |
AT5G47450
|
ATTIP2;3, ARABIDOPSIS THALIANA TONOPLAST INTRINSIC PROTEIN 2;3, DELTA-TIP3, DELTA-TONOPLAST INTRINSIC PROTEIN 3, TIP2;3, tonoplast intrinsic protein 2;3 |
-0.283 |
0.184 |
| 335 |
261839_at |
AT1G16040
|
unknown |
-0.282 |
0.169 |
| 336 |
266456_at |
AT2G22770
|
NAI1 |
-0.282 |
-0.627 |
| 337 |
260230_at |
AT1G74500
|
ATBS1, activation-tagged BRI1(brassinosteroid-insensitive 1)-suppressor 1, bHLH135, basic helix-loop-helix protein 135, BS1, activation-tagged BRI1(brassinosteroid-insensitive 1)-suppressor 1, PRE3, PACLOBUTRAZOL RESISTANCE 3, TMO7, TARGET OF MONOPTEROS 7 |
-0.280 |
-1.070 |
| 338 |
264725_at |
AT1G22885
|
unknown |
-0.279 |
0.115 |
| 339 |
261700_at |
AT1G32690
|
unknown |
-0.279 |
0.323 |
| 340 |
263046_at |
AT2G05380
|
GRP3S, glycine-rich protein 3 short isoform |
-0.277 |
0.766 |
| 341 |
254914_at |
AT4G11290
|
[Peroxidase superfamily protein] |
-0.276 |
-0.139 |
| 342 |
245550_at |
AT4G15330
|
CYP705A1, cytochrome P450, family 705, subfamily A, polypeptide 1 |
-0.275 |
-0.552 |
| 343 |
253317_at |
AT4G33960
|
unknown |
-0.274 |
-0.341 |
| 344 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
-0.274 |
-0.912 |
| 345 |
266168_at |
AT2G38870
|
[Serine protease inhibitor, potato inhibitor I-type family protein] |
-0.272 |
-0.055 |
| 346 |
257418_at |
AT1G30850
|
RSH4, root hair specific 4 |
-0.269 |
-0.935 |
| 347 |
260941_at |
AT1G44970
|
[Peroxidase superfamily protein] |
-0.269 |
0.067 |
| 348 |
263979_at |
AT2G42840
|
PDF1, protodermal factor 1 |
-0.268 |
1.548 |
| 349 |
247425_at |
AT5G62550
|
unknown |
-0.267 |
-0.162 |
| 350 |
261773_at |
AT1G76250
|
unknown |
-0.267 |
-1.253 |
| 351 |
258183_at |
AT3G21550
|
AtDMP2, Arabidopsis thaliana DUF679 domain membrane protein 2, DMP2, DUF679 domain membrane protein 2 |
-0.267 |
-0.049 |
| 352 |
256762_at |
AT3G25655
|
IDL1, inflorescence deficient in abscission (IDA)-like 1 |
-0.267 |
-0.198 |
| 353 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
-0.267 |
-0.079 |
| 354 |
262286_at |
AT1G68585
|
unknown |
-0.266 |
0.675 |
| 355 |
259211_at |
AT3G09020
|
[alpha 1,4-glycosyltransferase family protein] |
-0.265 |
-1.640 |
| 356 |
245566_at |
AT4G14610
|
[pseudogene] |
-0.265 |
-0.118 |
| 357 |
248732_at |
AT5G48070
|
ATXTH20, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 20, XTH20, xyloglucan endotransglucosylase/hydrolase 20 |
-0.264 |
-0.251 |
| 358 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
-0.263 |
0.285 |
| 359 |
248511_at |
AT5G50375
|
CPI1, cyclopropyl isomerase |
-0.261 |
1.199 |
| 360 |
256563_at |
AT3G29780
|
RALFL27, ralf-like 27 |
-0.261 |
-0.490 |
| 361 |
260151_at |
AT1G52910
|
unknown |
-0.260 |
0.204 |
| 362 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
-0.260 |
-0.205 |
| 363 |
251255_at |
AT3G62280
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.259 |
-0.351 |
| 364 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
-0.259 |
-0.429 |
| 365 |
257671_at |
AT3G20450
|
[B-cell receptor-associated protein 31-like ] |
-0.259 |
0.003 |
| 366 |
254294_at |
AT4G23070
|
ATRBL7, RHOMBOID-like protein 7, RBL7, RHOMBOID-like protein 7 |
-0.259 |
-0.472 |
| 367 |
258912_at |
AT3G06460
|
[GNS1/SUR4 membrane protein family] |
-0.258 |
0.389 |
| 368 |
266566_at |
AT2G24040
|
[Low temperature and salt responsive protein family] |
-0.258 |
1.110 |
| 369 |
258222_at |
AT3G15680
|
[Ran BP2/NZF zinc finger-like superfamily protein] |
-0.258 |
0.212 |
| 370 |
264577_at |
AT1G05260
|
RCI3, RARE COLD INDUCIBLE GENE 3, RCI3A |
-0.258 |
-1.500 |
| 371 |
262922_at |
AT1G79420
|
unknown |
-0.257 |
-0.382 |
| 372 |
250632_at |
AT5G07450
|
CYCP4;3, cyclin p4;3 |
-0.257 |
-1.070 |
| 373 |
260401_at |
AT1G69840
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
-0.257 |
0.001 |
| 374 |
246826_at |
AT5G26310
|
UGT72E3 |
-0.257 |
-1.382 |
| 375 |
259637_at |
AT1G52260
|
ATPDI3, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 3, ATPDIL1-5, PDI-like 1-5, PDI3, PROTEIN DISULFIDE ISOMERASE 3, PDIL1-5, PDI-like 1-5 |
-0.257 |
-0.813 |
| 376 |
264842_at |
AT1G03700
|
[Uncharacterised protein family (UPF0497)] |
-0.256 |
-0.093 |
| 377 |
263473_at |
AT2G31750
|
UGT74D1, UDP-glucosyl transferase 74D1 |
-0.256 |
0.233 |
| 378 |
258623_at |
AT3G02790
|
MBS1, METHYLENE BLUE SENSITIVITY 1 |
-0.255 |
0.942 |
| 379 |
253855_at |
AT4G28050
|
TET7, tetraspanin7 |
-0.255 |
0.901 |
| 380 |
258702_at |
AT3G09730
|
unknown |
-0.255 |
-0.376 |
| 381 |
262081_at |
AT1G59540
|
ZCF125 |
-0.255 |
0.005 |
| 382 |
258067_at |
AT3G25980
|
AtMAD2, MAD2, MITOTIC ARREST-DEFICIENT 2 |
-0.254 |
-0.080 |
| 383 |
250901_at |
AT5G03530
|
ATRAB, ATRAB ALPHA, ATRAB18B, ATRABC2A, ARABIDOPSIS THALIANA RAB GTPASE HOMOLOG C2A, RABC2A, RAB GTPase homolog C2A |
-0.253 |
-0.256 |
| 384 |
251405_at |
AT3G60330
|
AHA7, H(+)-ATPase 7, HA7, H(+)-ATPase 7 |
-0.253 |
-0.896 |
| 385 |
263398_at |
AT2G31680
|
AtRABA5d, RAB GTPase homolog A5D, RABA5d, RAB GTPase homolog A5D |
-0.253 |
-0.315 |
| 386 |
250391_at |
AT5G10850
|
[similar to nucleic acid binding / zinc ion binding [Arabidopsis thaliana] (TAIR:AT2G01050.1)] |
-0.252 |
-0.524 |
| 387 |
264377_at |
AT2G25060
|
AtENODL14, ENODL14, early nodulin-like protein 14 |
-0.251 |
0.674 |
| 388 |
252943_at |
AT4G39330
|
ATCAD9, CAD9, cinnamyl alcohol dehydrogenase 9 |
-0.251 |
1.453 |
| 389 |
266500_at |
AT2G06925
|
ATSPLA2-ALPHA, PHOSPHOLIPASE A2-ALPHA, PLA2-ALPHA |
-0.250 |
0.480 |
| 390 |
261220_at |
AT1G19970
|
[ER lumen protein retaining receptor family protein] |
-0.249 |
1.016 |
| 391 |
249184_at |
AT5G43020
|
[Leucine-rich repeat protein kinase family protein] |
-0.248 |
0.718 |
| 392 |
245359_at |
AT4G14430
|
ATECI2, ARABIDOPSIS THALIANA DELTA(3), DELTA(2)-ENOYL COA ISOMERASE 2, ECHIB, ENOYL-COA HYDRATASE/ISOMERASE B, ECI2, DELTA(3), DELTA(2)-ENOYL COA ISOMERASE 2, IBR10, indole-3-butyric acid response 10, PEC12 |
-0.247 |
0.550 |
| 393 |
260867_at |
AT1G43790
|
TED6, tracheary element differentiation-related 6 |
-0.247 |
0.021 |
| 394 |
253628_at |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
-0.246 |
-0.320 |
| 395 |
259479_at |
AT1G19020
|
unknown |
-0.246 |
-0.031 |
| 396 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
-0.246 |
-0.436 |
| 397 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
-0.245 |
-0.966 |
| 398 |
259688_at |
AT1G63120
|
ATRBL2, RHOMBOID-like 2, RBL2, RHOMBOID-like 2 |
-0.245 |
-0.015 |
| 399 |
260897_at |
AT1G29330
|
AERD2, ARABIDOPSIS ENDOPLASMIC RETICULUM RETENTION DEFECTIVE 2, ATERD2, ARABIDOPSIS THALIANA ENDOPLASMIC RETICULUM RETENTION DEFECTIVE 2, ERD2, ENDOPLASMIC RETICULUM RETENTION DEFECTIVE 2 |
-0.244 |
0.218 |
| 400 |
246581_at |
AT1G31760
|
[SWIB/MDM2 domain superfamily protein] |
-0.244 |
0.369 |
| 401 |
261727_at |
AT1G76090
|
SMT3, sterol methyltransferase 3 |
-0.244 |
-0.189 |
| 402 |
250517_at |
AT5G08260
|
scpl35, serine carboxypeptidase-like 35 |
-0.243 |
0.160 |
| 403 |
253626_at |
AT4G30640
|
[RNI-like superfamily protein] |
-0.243 |
-0.985 |
| 404 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
-0.243 |
-0.014 |
| 405 |
262230_at |
AT1G68560
|
ATXYL1, ALPHA-XYLOSIDASE 1, AXY3, altered xyloglucan 3, TRG1, thermoinhibition resistant germination 1, XYL1, alpha-xylosidase 1 |
-0.243 |
0.496 |
| 406 |
265663_at |
AT2G24290
|
unknown |
-0.242 |
0.638 |
| 407 |
267636_at |
AT2G42110
|
unknown |
-0.242 |
0.050 |
| 408 |
257755_at |
AT3G18760
|
[Translation elongation factor EF1B/ribosomal protein S6 family protein] |
-0.241 |
0.841 |
| 409 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
-0.241 |
-0.822 |
| 410 |
262457_at |
AT1G11200
|
unknown |
-0.241 |
0.132 |
| 411 |
247458_at |
AT5G62180
|
AtCXE20, carboxyesterase 20, CXE20, carboxyesterase 20 |
-0.240 |
-0.525 |
| 412 |
260034_at |
AT1G68810
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.238 |
0.194 |
| 413 |
253413_at |
AT4G33020
|
ATZIP9, ZIP9 |
-0.238 |
-0.447 |
| 414 |
266172_at |
AT2G39010
|
PIP2;6, PLASMA MEMBRANE INTRINSIC PROTEIN 2;6, PIP2E, plasma membrane intrinsic protein 2E |
-0.238 |
0.439 |
| 415 |
266336_at |
AT2G32270
|
ZIP3, zinc transporter 3 precursor |
-0.238 |
0.364 |
| 416 |
247970_at |
AT5G56720
|
c-NAD-MDH3, cytosolic-NAD-dependent malate dehydrogenase 3 |
-0.238 |
-0.166 |
| 417 |
261653_at |
AT1G01900
|
ATSBT1.1, SBTI1.1 |
-0.237 |
-0.230 |
| 418 |
245523_at |
AT4G15910
|
ATDI21, drought-induced 21, DI21, drought-induced 21 |
-0.237 |
1.251 |
| 419 |
261167_at |
AT1G04980
|
ATPDI10, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 10, ATPDIL2-2, PDI-like 2-2, PDI10, PROTEIN DISULFIDE ISOMERASE, PDIL2-2, PDI-like 2-2 |
-0.237 |
0.348 |
| 420 |
266244_at |
AT2G27740
|
unknown |
-0.237 |
-0.113 |
| 421 |
246143_at |
AT5G19980
|
GONST4, golgi nucleotide sugar transporter 4 |
-0.236 |
0.171 |
| 422 |
267344_at |
AT2G44230
|
unknown |
-0.236 |
-0.478 |
| 423 |
265823_at |
AT2G35760
|
[Uncharacterised protein family (UPF0497)] |
-0.235 |
0.012 |
| 424 |
258584_at |
AT3G04090
|
SIP1;1, SIP1A, small and basic intrinsic protein 1A |
-0.235 |
0.416 |
| 425 |
260585_at |
AT2G43650
|
EMB2777, EMBRYO DEFECTIVE 2777 |
-0.235 |
1.100 |
| 426 |
252711_at |
AT3G43720
|
LTPG2, glycosylphosphatidylinositol-anchored lipid protein transfer 2 |
-0.235 |
1.288 |
| 427 |
258599_at |
AT3G04520
|
THA2, threonine aldolase 2 |
-0.234 |
0.471 |
| 428 |
255885_at |
AT1G20330
|
CVP1, COTYLEDON VASCULAR PATTERN 1, FRL1, FRILL1, SMT2, sterol methyltransferase 2 |
-0.233 |
1.302 |
| 429 |
247741_at |
AT5G58960
|
GIL1, GRAVITROPIC IN THE LIGHT |
-0.233 |
0.064 |
| 430 |
262408_at |
AT1G34750
|
[Protein phosphatase 2C family protein] |
-0.233 |
0.291 |
| 431 |
257467_at |
AT1G31320
|
LBD4, LOB domain-containing protein 4 |
-0.232 |
-0.370 |
| 432 |
251297_at |
AT3G62020
|
GLP10, germin-like protein 10 |
-0.231 |
-1.022 |
| 433 |
253910_at |
AT4G27290
|
[S-locus lectin protein kinase family protein] |
-0.230 |
-1.463 |
| 434 |
252906_at |
AT4G39640
|
GGT1, gamma-glutamyl transpeptidase 1 |
-0.230 |
0.596 |
| 435 |
249675_at |
AT5G35940
|
[Mannose-binding lectin superfamily protein] |
-0.230 |
-0.595 |