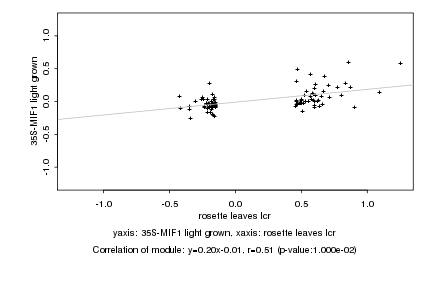
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
rosette leaves lcr |
Log2 signal ratio
35S-MIF1 light grown |
| 1 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
1.249 |
0.593 |
| 2 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
1.089 |
0.139 |
| 3 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
0.898 |
-0.078 |
| 4 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
0.867 |
0.228 |
| 5 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.853 |
0.605 |
| 6 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
0.831 |
0.289 |
| 7 |
251065_at |
AT5G01870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.800 |
0.100 |
| 8 |
250365_at |
AT5G11410
|
[Protein kinase superfamily protein] |
0.774 |
0.223 |
| 9 |
267008_at |
AT2G39350
|
ABCG1, ATP-binding cassette G1 |
0.707 |
0.076 |
| 10 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.702 |
0.250 |
| 11 |
258209_at |
AT3G14060
|
unknown |
0.668 |
0.385 |
| 12 |
248611_at |
AT5G49520
|
ATWRKY48, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 48, WRKY48, WRKY DNA-binding protein 48 |
0.661 |
0.153 |
| 13 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.658 |
-0.042 |
| 14 |
254652_at |
AT4G18170
|
ATWRKY28, WRKY28, WRKY DNA-binding protein 28 |
0.646 |
0.090 |
| 15 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.637 |
-0.069 |
| 16 |
246831_at |
AT5G26340
|
ATSTP13, SUGAR TRANSPORT PROTEIN 13, MSS1, STP13, SUGAR TRANSPORT PROTEIN 13 |
0.624 |
0.017 |
| 17 |
255250_at |
AT4G05100
|
AtMYB74, myb domain protein 74, MYB74, myb domain protein 74 |
0.615 |
0.005 |
| 18 |
266246_at |
AT2G27690
|
CYP94C1, cytochrome P450, family 94, subfamily C, polypeptide 1 |
0.606 |
0.097 |
| 19 |
262887_at |
AT1G14780
|
[MAC/Perforin domain-containing protein] |
0.603 |
0.272 |
| 20 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
0.599 |
0.199 |
| 21 |
248218_at |
AT5G53710
|
unknown |
0.598 |
-0.055 |
| 22 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.594 |
0.001 |
| 23 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.593 |
-0.085 |
| 24 |
264580_at |
AT1G05340
|
unknown |
0.592 |
0.114 |
| 25 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.590 |
0.027 |
| 26 |
247013_at |
AT5G67480
|
ATBT4, BT4, BTB and TAZ domain protein 4 |
0.581 |
0.128 |
| 27 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
0.572 |
0.034 |
| 28 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.564 |
0.425 |
| 29 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
0.563 |
0.078 |
| 30 |
252511_at |
AT3G46280
|
[protein kinase-related] |
0.552 |
0.009 |
| 31 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
0.536 |
0.153 |
| 32 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
0.527 |
0.008 |
| 33 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
0.520 |
0.098 |
| 34 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
0.509 |
-0.028 |
| 35 |
248163_at |
AT5G54510
|
DFL1, DWARF IN LIGHT 1, GH3.6, Gretchen Hagen3.6 |
0.505 |
-0.137 |
| 36 |
261088_at |
AT1G07590
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.501 |
0.018 |
| 37 |
261410_at |
AT1G07610
|
MT1C, metallothionein 1C |
0.499 |
0.042 |
| 38 |
253872_at |
AT4G27410
|
ANAC072, Arabidopsis NAC domain containing protein 72, RD26, RESPONSIVE TO DESICCATION 26 |
0.498 |
-0.025 |
| 39 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
0.493 |
-0.014 |
| 40 |
252487_at |
AT3G46660
|
UGT76E12, UDP-glucosyl transferase 76E12 |
0.488 |
-0.018 |
| 41 |
259445_at |
AT1G02400
|
ATGA2OX4, Arabidopsis thaliana gibberellin 2-oxidase 4, ATGA2OX6, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 6, DTA1, DOWNSTREAM TARGET OF AGL15 1, GA2OX6, gibberellin 2-oxidase 6 |
0.485 |
-0.017 |
| 42 |
256933_at |
AT3G22600
|
LTPG5, glycosylphosphatidylinositol-anchored lipid protein transfer 5 |
0.468 |
-0.020 |
| 43 |
256114_at |
AT1G16850
|
unknown |
0.467 |
-0.044 |
| 44 |
264960_at |
AT1G76930
|
ATEXT1, EXTENSIN 1, ATEXT4, extensin 4, EXT1, extensin 1, EXT4, extensin 4, ORG5, OBP3-RESPONSIVE GENE 5 |
0.464 |
0.501 |
| 45 |
247492_at |
AT5G61890
|
[Integrase-type DNA-binding superfamily protein] |
0.463 |
-0.035 |
| 46 |
254314_at |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
0.461 |
0.317 |
| 47 |
250826_at |
AT5G05220
|
unknown |
0.460 |
0.015 |
| 48 |
262947_at |
AT1G75750
|
GASA1, GAST1 protein homolog 1 |
0.458 |
0.020 |
| 49 |
249626_at |
AT5G37540
|
[Eukaryotic aspartyl protease family protein] |
0.455 |
-0.074 |
| 50 |
250944_at |
AT5G03380
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.428 |
0.089 |
| 51 |
266921_at |
AT2G45970
|
CYP86A8, cytochrome P450, family 86, subfamily A, polypeptide 8, LCR, LACERATA |
-0.421 |
-0.105 |
| 52 |
246535_at |
AT5G15900
|
TBL19, TRICHOME BIREFRINGENCE-LIKE 19 |
-0.356 |
-0.064 |
| 53 |
265698_at |
AT2G32160
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.350 |
-0.108 |
| 54 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.344 |
-0.248 |
| 55 |
263480_at |
AT2G04032
|
ZIP7, zinc transporter 7 precursor |
-0.309 |
0.001 |
| 56 |
251826_at |
AT3G55110
|
ABCG18, ATP-binding cassette G18 |
-0.263 |
0.033 |
| 57 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
-0.256 |
0.074 |
| 58 |
246769_at |
AT5G27440
|
unknown |
-0.243 |
0.034 |
| 59 |
258932_at |
AT3G10150
|
ATPAP16, PAP16, purple acid phosphatase 16 |
-0.242 |
-0.063 |
| 60 |
267078_at |
AT2G40960
|
[Single-stranded nucleic acid binding R3H protein] |
-0.237 |
-0.089 |
| 61 |
246235_at |
AT4G36830
|
HOS3-1 |
-0.222 |
-0.022 |
| 62 |
251509_at |
AT3G59010
|
PME35, pectin methylesterase 35, PME61, pectin methylesterase 61 |
-0.220 |
-0.030 |
| 63 |
263073_at |
AT2G17500
|
[Auxin efflux carrier family protein] |
-0.217 |
0.035 |
| 64 |
266321_at |
AT2G46660
|
CYP78A6, cytochrome P450, family 78, subfamily A, polypeptide 6, EOD3, enhancer of da1-1 |
-0.216 |
-0.092 |
| 65 |
249134_at |
AT5G43150
|
unknown |
-0.215 |
-0.152 |
| 66 |
253518_at |
AT4G31400
|
AtCTF7, CTF7, CHROMOSOME TRANSMISSION FIDELITY 7, ECO1 |
-0.214 |
-0.036 |
| 67 |
250372_at |
AT5G11460
|
unknown |
-0.205 |
-0.018 |
| 68 |
249567_at |
AT5G38020
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.201 |
0.276 |
| 69 |
248078_at |
AT5G55780
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.197 |
-0.083 |
| 70 |
255450_at |
AT4G02850
|
[phenazine biosynthesis PhzC/PhzF family protein] |
-0.197 |
-0.067 |
| 71 |
267402_at |
AT2G26180
|
IQD6, IQ-domain 6 |
-0.197 |
-0.037 |
| 72 |
263042_at |
AT1G23340
|
unknown |
-0.197 |
-0.044 |
| 73 |
247118_at |
AT5G65890
|
ACR1, ACT domain repeat 1 |
-0.190 |
-0.156 |
| 74 |
248901_at |
AT5G46410
|
SSP4, SCP1-like small phosphatase 4 |
-0.189 |
0.014 |
| 75 |
259888_at |
AT1G76350
|
NLP5, NIN-like protein 5 |
-0.186 |
-0.044 |
| 76 |
261084_at |
AT1G07440
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.185 |
-0.080 |
| 77 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.184 |
-0.120 |
| 78 |
262312_at |
AT1G70830
|
MLP28, MLP-like protein 28 |
-0.181 |
0.114 |
| 79 |
251733_at |
AT3G56240
|
CCH, copper chaperone |
-0.180 |
-0.190 |
| 80 |
252594_at |
AT3G45680
|
[Major facilitator superfamily protein] |
-0.180 |
-0.087 |
| 81 |
262907_at |
AT1G59720
|
CRR28, CHLORORESPIRATORY REDUCTION28 |
-0.179 |
-0.049 |
| 82 |
263887_at |
AT2G36980
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.176 |
-0.023 |
| 83 |
246624_at |
AT1G48910
|
YUC10, YUCCA 10 |
-0.173 |
-0.043 |
| 84 |
258561_at |
AT3G05960
|
ATSTP6, SUGAR TRANSPORTER 6, STP6, sugar transporter 6 |
-0.170 |
-0.051 |
| 85 |
264712_at |
AT1G09810
|
ECT11, evolutionarily conserved C-terminal region 11 |
-0.168 |
0.043 |
| 86 |
251968_at |
AT3G53100
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.167 |
-0.199 |
| 87 |
257459_at |
AT1G24040
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.166 |
0.067 |
| 88 |
265838_at |
AT2G14550
|
[pseudogene] |
-0.165 |
-0.018 |
| 89 |
254153_at |
AT4G24450
|
ATGWD2, GWD3, PWD, phosphoglucan, water dikinase |
-0.165 |
-0.213 |
| 90 |
245147_at |
AT2G45280
|
ATRAD51C, RAS associated with diabetes protein 51C, RAD51C, RAS associated with diabetes protein 51C |
-0.160 |
0.042 |
| 91 |
257528_at |
AT3G02125
|
unknown |
-0.159 |
-0.015 |
| 92 |
251870_at |
AT3G54510
|
[Early-responsive to dehydration stress protein (ERD4)] |
-0.158 |
-0.060 |
| 93 |
248494_at |
AT5G50770
|
AtHSD6, hydroxysteroid dehydrogenase 6, HSD6, hydroxysteroid dehydrogenase 6 |
-0.156 |
-0.066 |
| 94 |
256646_at |
AT3G13590
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.154 |
-0.064 |
| 95 |
246023_at |
AT5G21120
|
EIL2, ETHYLENE-INSENSITIVE3-like 2 |
-0.154 |
-0.032 |
| 96 |
261200_at |
AT1G12960
|
[Ribosomal protein L18e/L15 superfamily protein] |
-0.154 |
-0.008 |
| 97 |
247243_at |
AT5G64700
|
UMAMIT21, Usually multiple acids move in and out Transporters 21 |
-0.153 |
-0.079 |