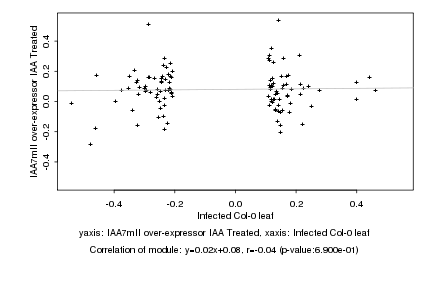
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Infected Col-0 leaf |
Log2 signal ratio
IAA7mII over-expressor IAA Treated |
| 1 |
248333_at |
AT5G52390
|
[PAR1 protein] |
0.461 |
0.074 |
| 2 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
0.440 |
0.164 |
| 3 |
250561_at |
AT5G08030
|
AtGDPD6, GDPD6, glycerophosphodiester phosphodiesterase 6 |
0.399 |
0.015 |
| 4 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.396 |
0.127 |
| 5 |
257017_at |
AT3G19620
|
[Glycosyl hydrolase family protein] |
0.274 |
0.074 |
| 6 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.250 |
-0.028 |
| 7 |
254024_at |
AT4G25780
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.239 |
0.104 |
| 8 |
258890_at |
AT3G05690
|
ATHAP2B, HEME ACTIVATOR PROTEIN (YEAST) HOMOLOG 2B, HAP2B, HEME ACTIVATOR PROTEIN (YEAST) HOMOLOG 2B, NF-YA2, nuclear factor Y, subunit A2, UNE8, UNFERTILIZED EMBRYO SAC 8 |
0.223 |
0.091 |
| 9 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.219 |
-0.147 |
| 10 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
0.214 |
0.051 |
| 11 |
247224_at |
AT5G65080
|
AGL68, AGAMOUS-like 68, MAF5, MADS AFFECTING FLOWERING 5 |
0.212 |
0.118 |
| 12 |
250688_at |
AT5G06510
|
NF-YA10, nuclear factor Y, subunit A10 |
0.210 |
0.305 |
| 13 |
258957_at |
AT3G01420
|
ALPHA-DOX1, alpha-dioxygenase 1, DIOX1, DOX1, PADOX-1, plant alpha dioxygenase 1 |
0.183 |
0.086 |
| 14 |
267181_at |
AT2G37760
|
AKR4C8, Aldo-keto reductase family 4 member C8 |
0.181 |
-0.007 |
| 15 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
0.176 |
-0.067 |
| 16 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.172 |
0.176 |
| 17 |
247704_at |
AT5G59510
|
DVL18, DEVIL 18, RTFL5, ROTUNDIFOLIA like 5 |
0.171 |
0.034 |
| 18 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.169 |
0.044 |
| 19 |
262357_at |
AT1G73040
|
[Mannose-binding lectin superfamily protein] |
0.167 |
0.171 |
| 20 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.166 |
0.118 |
| 21 |
265161_at |
AT1G30900
|
BP80-3;3, binding protein of 80 kDa 3;3, VSR3;3, VACUOLAR SORTING RECEPTOR 3;3, VSR6, VACUOLAR SORTING RECEPTOR 6 |
0.158 |
0.108 |
| 22 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
0.158 |
0.288 |
| 23 |
264635_at |
AT1G65500
|
unknown |
0.154 |
0.091 |
| 24 |
246075_at |
AT5G20410
|
ATMGD2, ARABIDOPSIS THALIANA MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE 2, MGD2, monogalactosyldiacylglycerol synthase 2 |
0.152 |
-0.057 |
| 25 |
257896_at |
AT3G16920
|
ATCTL2, CTL2, chitinase-like protein 2 |
0.150 |
0.171 |
| 26 |
254759_at |
AT4G13180
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.148 |
-0.157 |
| 27 |
256100_at |
AT1G13750
|
[Purple acid phosphatases superfamily protein] |
0.147 |
-0.070 |
| 28 |
248330_at |
AT5G52810
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.147 |
-0.201 |
| 29 |
255543_at |
AT4G01870
|
[tolB protein-related] |
0.143 |
0.016 |
| 30 |
259133_at |
AT3G05400
|
[Major facilitator superfamily protein] |
0.141 |
0.537 |
| 31 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
0.141 |
-0.066 |
| 32 |
262378_at |
AT1G72830
|
ATHAP2C, HAP2C, NF-YA3, nuclear factor Y, subunit A3 |
0.140 |
-0.024 |
| 33 |
261399_at |
AT1G79620
|
[Leucine-rich repeat protein kinase family protein] |
0.136 |
0.055 |
| 34 |
264741_at |
AT1G62290
|
[Saposin-like aspartyl protease family protein] |
0.136 |
-0.127 |
| 35 |
260771_at |
AT1G49160
|
WNK7 |
0.133 |
0.072 |
| 36 |
254027_at |
AT4G25835
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.131 |
-0.058 |
| 37 |
266296_at |
AT2G29420
|
ATGSTU7, glutathione S-transferase tau 7, GST25, GLUTATHIONE S-TRANSFERASE 25, GSTU7, glutathione S-transferase tau 7 |
0.130 |
-0.047 |
| 38 |
245912_at |
AT5G19600
|
SULTR3;5, sulfate transporter 3;5 |
0.129 |
0.052 |
| 39 |
247223_at |
AT5G65070
|
AGL69, AGAMOUS-like 69, FCL4, MAF4, MADS AFFECTING FLOWERING 4 |
0.127 |
0.018 |
| 40 |
254618_at |
AT4G18780
|
ATCESA8, CELLULOSE SYNTHASE 8, CESA8, CELLULOSE SYNTHASE 8, IRX1, IRREGULAR XYLEM 1, LEW2, LEAF WILTING 2 |
0.124 |
0.264 |
| 41 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
0.124 |
0.125 |
| 42 |
250992_at |
AT5G02260
|
ATEXP9, ATEXPA9, expansin A9, ATHEXP ALPHA 1.10, EXP9, EXPA9, expansin A9 |
0.124 |
0.015 |
| 43 |
246425_at |
AT5G17420
|
ATCESA7, CESA7, CELLULOSE SYNTHASE CATALYTIC SUBUNIT 7, IRX3, IRREGULAR XYLEM 3, MUR10, MURUS 10 |
0.122 |
0.106 |
| 44 |
260684_at |
AT1G17590
|
NF-YA8, nuclear factor Y, subunit A8 |
0.121 |
0.157 |
| 45 |
265463_at |
AT2G37090
|
IRX9, IRREGULAR XYLEM 9 |
0.121 |
-0.006 |
| 46 |
257824_at |
AT3G25290
|
[Auxin-responsive family protein] |
0.118 |
0.355 |
| 47 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
0.118 |
0.004 |
| 48 |
259281_at |
AT3G01140
|
AtMYB106, myb domain protein 106, MYB106, myb domain protein 106, NOK, NOECK |
0.116 |
0.108 |
| 49 |
249397_at |
AT5G40230
|
UMAMIT37, Usually multiple acids move in and out Transporters 37 |
0.116 |
0.019 |
| 50 |
249070_at |
AT5G44030
|
CESA4, cellulose synthase A4, IRX5, IRREGULAR XYLEM 5, NWS2 |
0.115 |
0.141 |
| 51 |
248121_at |
AT5G54690
|
GAUT12, galacturonosyltransferase 12, IRX8, IRREGULAR XYLEM 8, LGT6 |
0.114 |
0.095 |
| 52 |
250820_at |
AT5G05160
|
RUL1, REDUCED IN LATERAL GROWTH1 |
0.113 |
0.081 |
| 53 |
251293_at |
AT3G61930
|
unknown |
0.112 |
0.309 |
| 54 |
257151_at |
AT3G27200
|
[Cupredoxin superfamily protein] |
0.112 |
0.109 |
| 55 |
260396_at |
AT1G69720
|
HO3, heme oxygenase 3 |
0.111 |
0.272 |
| 56 |
266313_at |
AT2G26980
|
CIPK3, CBL-interacting protein kinase 3, SnRK3.17, SNF1-RELATED PROTEIN KINASE 3.17 |
0.110 |
-0.024 |
| 57 |
248670_at |
AT5G48740
|
[Leucine-rich repeat protein kinase family protein] |
0.108 |
0.035 |
| 58 |
267094_at |
AT2G38080
|
ATLMCO4, ARABIDOPSIS LACCASE-LIKE MULTICOPPER OXIDASE 4, IRX12, IRREGULAR XYLEM 12, LAC4, LACCASE 4, LMCO4, LACCASE-LIKE MULTICOPPER OXIDASE 4 |
0.106 |
0.286 |
| 59 |
249599_at |
AT5G37990
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.543 |
-0.012 |
| 60 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
-0.478 |
-0.282 |
| 61 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.463 |
-0.176 |
| 62 |
264960_at |
AT1G76930
|
ATEXT1, EXTENSIN 1, ATEXT4, extensin 4, EXT1, extensin 1, EXT4, extensin 4, ORG5, OBP3-RESPONSIVE GENE 5 |
-0.459 |
0.173 |
| 63 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
-0.397 |
0.006 |
| 64 |
258791_at |
AT3G04720
|
AtPR4, HEL, HEVEIN-LIKE, PR-4, PR4, pathogenesis-related 4 |
-0.377 |
0.075 |
| 65 |
255725_at |
AT1G25540
|
MED25, mediator 25, PFT1, PHYTOCHROME AND FLOWERING TIME 1 |
-0.355 |
0.089 |
| 66 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
-0.352 |
0.169 |
| 67 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
-0.340 |
-0.056 |
| 68 |
253832_at |
AT4G27654
|
unknown |
-0.333 |
0.208 |
| 69 |
265917_at |
AT2G15080
|
AtRLP19, receptor like protein 19, RLP19, receptor like protein 19 |
-0.327 |
0.130 |
| 70 |
259429_at |
AT1G01600
|
CYP86A4, cytochrome P450, family 86, subfamily A, polypeptide 4 |
-0.326 |
0.141 |
| 71 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
-0.324 |
-0.153 |
| 72 |
249726_at |
AT5G35480
|
unknown |
-0.321 |
0.050 |
| 73 |
256569_at |
AT3G19550
|
unknown |
-0.319 |
0.099 |
| 74 |
249895_at |
AT5G22500
|
FAR1, fatty acid reductase 1 |
-0.302 |
0.092 |
| 75 |
264574_at |
AT1G05300
|
ZIP5, zinc transporter 5 precursor |
-0.297 |
0.068 |
| 76 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
-0.297 |
0.102 |
| 77 |
256527_at |
AT1G66100
|
[Plant thionin] |
-0.294 |
0.075 |
| 78 |
254907_at |
AT4G11190
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.289 |
0.164 |
| 79 |
265539_at |
AT2G15830
|
unknown |
-0.287 |
0.516 |
| 80 |
246000_at |
AT5G20820
|
SAUR76, SMALL AUXIN UPREGULATED RNA 76 |
-0.285 |
0.160 |
| 81 |
252520_at |
AT3G46370
|
[Leucine-rich repeat protein kinase family protein] |
-0.283 |
0.062 |
| 82 |
246687_at |
AT5G33370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.267 |
0.159 |
| 83 |
258897_at |
AT3G05730
|
[LOCATED IN: endomembrane system] |
-0.263 |
0.029 |
| 84 |
265656_at |
AT2G13820
|
AtXYP2, XYP2, xylogen protein 2 |
-0.260 |
0.085 |
| 85 |
257288_at |
AT3G29670
|
PMAT2, phenolic glucoside malonyltransferase 2 |
-0.260 |
0.053 |
| 86 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
-0.255 |
-0.103 |
| 87 |
254150_at |
AT4G24350
|
[Phosphorylase superfamily protein] |
-0.253 |
0.004 |
| 88 |
253911_at |
AT4G27300
|
[S-locus lectin protein kinase family protein] |
-0.249 |
0.070 |
| 89 |
246949_at |
AT5G25140
|
CYP71B13, cytochrome P450, family 71, subfamily B, polypeptide 13 |
-0.248 |
-0.043 |
| 90 |
250043_at |
AT5G18430
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.246 |
0.136 |
| 91 |
255786_at |
AT1G19670
|
ATCLH1, chlorophyllase 1, ATHCOR1, CORONATINE-INDUCED PROTEIN 1, CLH1, chlorophyllase 1, CORI1, CORONATINE-INDUCED PROTEIN 1 |
-0.245 |
0.127 |
| 92 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
-0.245 |
0.158 |
| 93 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
-0.243 |
0.168 |
| 94 |
266222_at |
AT2G28780
|
unknown |
-0.239 |
-0.097 |
| 95 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
-0.238 |
0.239 |
| 96 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.236 |
0.285 |
| 97 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
-0.236 |
0.023 |
| 98 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
-0.236 |
-0.021 |
| 99 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
-0.234 |
-0.182 |
| 100 |
265441_at |
AT2G20870
|
[cell wall protein precursor, putative] |
-0.231 |
0.151 |
| 101 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
-0.231 |
0.079 |
| 102 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
-0.229 |
0.230 |
| 103 |
266415_at |
AT2G38530
|
cdf3, cell growth defect factor-3, LP2, LTP2, lipid transfer protein 2 |
-0.227 |
-0.142 |
| 104 |
254697_at |
AT4G17970
|
ALMT12, aluminum-activated, malate transporter 12, ATALMT12, QUAC1, quick-activating anion channel 1 |
-0.223 |
0.180 |
| 105 |
260241_at |
AT1G63710
|
CYP86A7, cytochrome P450, family 86, subfamily A, polypeptide 7 |
-0.221 |
0.079 |
| 106 |
249567_at |
AT5G38020
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.220 |
0.092 |
| 107 |
261375_at |
AT1G53160
|
FTM6, FLORAL TRANSITION AT THE MERISTEM6, SPL4, squamosa promoter binding protein-like 4 |
-0.219 |
0.131 |
| 108 |
258419_at |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.217 |
0.086 |
| 109 |
248073_at |
AT5G55720
|
[Pectin lyase-like superfamily protein] |
-0.215 |
0.168 |
| 110 |
267509_at |
AT2G45660
|
AGL20, AGAMOUS-like 20, ATSOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1, SOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1 |
-0.215 |
0.253 |
| 111 |
251617_at |
AT3G58000
|
[VQ motif-containing protein] |
-0.214 |
0.055 |
| 112 |
247882_at |
AT5G57785
|
unknown |
-0.212 |
0.062 |
| 113 |
249727_at |
AT5G35490
|
ATMRU1, ARABIDOPSIS MTO 1 RESPONDING UP 1, MRU1, MTO 1 RESPONDING UP 1 |
-0.212 |
0.160 |
| 114 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
-0.209 |
0.203 |
| 115 |
263480_at |
AT2G04032
|
ZIP7, zinc transporter 7 precursor |
-0.208 |
0.035 |