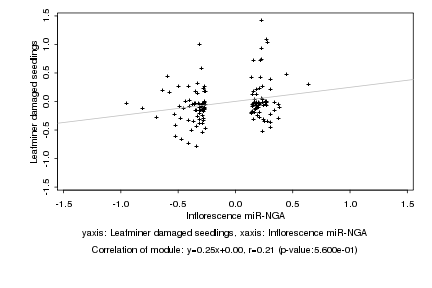
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Inflorescence miR-NGA |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
246437_at |
AT5G17540
|
[HXXXD-type acyl-transferase family protein] |
0.634 |
0.309 |
| 2 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.444 |
0.487 |
| 3 |
254037_at |
AT4G25760
|
ATGDU2, glutamine dumper 2, GDU2, glutamine dumper 2 |
0.376 |
-0.090 |
| 4 |
248867_at |
AT5G46830
|
ATNIG1, NACL-INDUCIBLE GENE 1, NIG1, NACL-inducible gene 1 |
0.368 |
-0.294 |
| 5 |
261576_at |
AT1G01070
|
UMAMIT28, Usually multiple acids move in and out Transporters 28 |
0.368 |
-0.044 |
| 6 |
251298_at |
AT3G62040
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.340 |
-0.142 |
| 7 |
251825_at |
AT3G55100
|
ABCG17, ATP-binding cassette G17 |
0.337 |
-0.009 |
| 8 |
261991_at |
AT1G33700
|
[Beta-glucosidase, GBA2 type family protein] |
0.303 |
-0.211 |
| 9 |
248772_at |
AT5G47800
|
[Phototropic-responsive NPH3 family protein] |
0.302 |
-0.438 |
| 10 |
257900_at |
AT3G28420
|
[Putative membrane lipoprotein] |
0.301 |
-0.363 |
| 11 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
0.301 |
0.215 |
| 12 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
0.298 |
0.394 |
| 13 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
0.278 |
1.035 |
| 14 |
262196_at |
AT1G77870
|
MUB5, membrane-anchored ubiquitin-fold protein 5 precursor |
0.276 |
-0.349 |
| 15 |
258267_at |
AT3G15870
|
[Fatty acid desaturase family protein] |
0.268 |
0.012 |
| 16 |
249955_at |
AT5G18840
|
[Major facilitator superfamily protein] |
0.267 |
-0.053 |
| 17 |
259793_at |
AT1G64380
|
[Integrase-type DNA-binding superfamily protein] |
0.266 |
1.093 |
| 18 |
256319_at |
AT1G35910
|
TPPD, trehalose-6-phosphate phosphatase D |
0.265 |
-0.001 |
| 19 |
254907_at |
AT4G11190
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.260 |
-0.004 |
| 20 |
266353_at |
AT2G01520
|
MLP328, MLP-like protein 328, ZCE1, (Zusammen-CA)-enhanced 1 |
0.257 |
-0.036 |
| 21 |
257570_at |
AT3G13662
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.256 |
0.003 |
| 22 |
255578_at |
AT4G01450
|
UMAMIT30, Usually multiple acids move in and out Transporters 30 |
0.247 |
-0.345 |
| 23 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
0.238 |
-0.307 |
| 24 |
257115_at |
AT3G20150
|
[Kinesin motor family protein] |
0.237 |
-0.068 |
| 25 |
267571_at |
AT2G30650
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
0.234 |
0.050 |
| 26 |
247288_at |
AT5G64330
|
JK218, NPH3, NON-PHOTOTROPIC HYPOCOTYL 3, RPT3, ROOT PHOTOTROPISM 3 |
0.231 |
-0.510 |
| 27 |
264363_at |
AT1G03170
|
FAF2, FANTASTIC FOUR 2, FTM5, FLORAL TRANSITION AT THE MERISTEM5 |
0.231 |
-0.001 |
| 28 |
259409_at |
AT1G13330
|
AHP2, Arabidopsis Hop2 homolog |
0.229 |
-0.015 |
| 29 |
251964_at |
AT3G53370
|
[S1FA-like DNA-binding protein] |
0.228 |
0.274 |
| 30 |
260263_at |
AT1G68480
|
JAG, JAGGED |
0.224 |
0.059 |
| 31 |
252076_at |
AT3G51660
|
[Tautomerase/MIF superfamily protein] |
0.223 |
0.935 |
| 32 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.222 |
1.437 |
| 33 |
245244_at |
AT1G44350
|
ILL6, IAA-leucine resistant (ILR)-like gene 6 |
0.220 |
0.747 |
| 34 |
259609_at |
AT1G52410
|
AtTSA1, TSA1, TSK-associating protein 1 |
0.211 |
0.727 |
| 35 |
252944_at |
AT4G39320
|
[microtubule-associated protein-related] |
0.211 |
0.431 |
| 36 |
260059_at |
AT1G78090
|
ATTPPB, Arabidopsis thaliana trehalose-6-phosphate phosphatase B, TPPB, trehalose-6-phosphate phosphatase B |
0.207 |
-0.178 |
| 37 |
248371_at |
AT5G51810
|
AT2353, ATGA20OX2, GA20OX2, gibberellin 20 oxidase 2 |
0.207 |
-0.043 |
| 38 |
245204_at |
AT5G12270
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.206 |
-0.023 |
| 39 |
264071_at |
AT2G27920
|
SCPL51, serine carboxypeptidase-like 51 |
0.205 |
-0.273 |
| 40 |
248770_at |
AT5G47740
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.204 |
0.238 |
| 41 |
254488_at |
AT4G20790
|
[Leucine-rich repeat protein kinase family protein] |
0.200 |
-0.013 |
| 42 |
258562_at |
AT3G05980
|
unknown |
0.198 |
-0.103 |
| 43 |
251555_at |
AT3G58780
|
AGL1, AGAMOUS-like 1, SHP1, SHATTERPROOF 1 |
0.194 |
-0.060 |
| 44 |
250504_at |
AT5G09840
|
[Putative endonuclease or glycosyl hydrolase] |
0.188 |
-0.104 |
| 45 |
245029_at |
AT2G26580
|
YAB5, YABBY5 |
0.184 |
-0.242 |
| 46 |
267298_at |
AT2G23760
|
BLH4, BEL1-like homeodomain 4, SAW2, SAWTOOTH 2 |
0.180 |
-0.069 |
| 47 |
258741_at |
AT3G05790
|
LON4, lon protease 4 |
0.178 |
-0.019 |
| 48 |
266139_at |
AT2G28085
|
SAUR42, SMALL AUXIN UPREGULATED RNA 42 |
0.178 |
0.215 |
| 49 |
254022_at |
AT4G25750
|
ABCG4, ATP-binding cassette G4 |
0.177 |
-0.004 |
| 50 |
245998_at |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
0.176 |
0.139 |
| 51 |
248280_at |
AT5G52950
|
unknown |
0.175 |
-0.120 |
| 52 |
249495_at |
AT5G39100
|
GLP6, germin-like protein 6 |
0.171 |
-0.020 |
| 53 |
246487_at |
AT5G16030
|
unknown |
0.169 |
-0.135 |
| 54 |
247068_at |
AT5G66800
|
unknown |
0.166 |
-0.006 |
| 55 |
252872_at |
AT4G40010
|
SNRK2-7, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-7, SNRK2.7, SNF1-related protein kinase 2.7, SRK2F |
0.166 |
-0.002 |
| 56 |
245108_at |
AT2G41510
|
ATCKX1, CYTOKININ OXIDASE/DEHYDROGENASE 1, CKX1, cytokinin oxidase/dehydrogenase 1 |
0.159 |
0.046 |
| 57 |
256652_at |
AT3G18850
|
LPAT5, lysophosphatidyl acyltransferase 5 |
0.158 |
-0.189 |
| 58 |
257376_at |
AT2G32350
|
[Ubiquitin-like superfamily protein] |
0.157 |
-0.027 |
| 59 |
261636_at |
AT1G50110
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
0.155 |
-0.307 |
| 60 |
263047_at |
AT2G17630
|
PSAT2, phosphoserine aminotransferase 2 |
0.154 |
-0.062 |
| 61 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
0.150 |
0.184 |
| 62 |
248395_at |
AT5G52120
|
AtPP2-A14, phloem protein 2-A14, PP2-A14, phloem protein 2-A14 |
0.149 |
0.732 |
| 63 |
262905_at |
AT1G59730
|
ATH7, thioredoxin H-type 7, TH7, thioredoxin H-type 7 |
0.148 |
-0.044 |
| 64 |
259014_at |
AT3G07320
|
[O-Glycosyl hydrolases family 17 protein] |
0.146 |
0.137 |
| 65 |
267402_at |
AT2G26180
|
IQD6, IQ-domain 6 |
0.142 |
-0.158 |
| 66 |
263441_at |
AT2G28620
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.141 |
-0.084 |
| 67 |
260618_at |
AT1G53230
|
TCP3, TEOSINTE BRANCHED 1, cycloidea and PCF transcription factor 3 |
0.140 |
-0.189 |
| 68 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
0.140 |
0.437 |
| 69 |
250201_at |
AT5G14230
|
unknown |
0.139 |
-0.198 |
| 70 |
256603_at |
AT3G28270
|
unknown |
-0.952 |
-0.020 |
| 71 |
264636_at |
AT1G65490
|
unknown |
-0.815 |
-0.116 |
| 72 |
249774_at |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
-0.697 |
-0.279 |
| 73 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.645 |
0.207 |
| 74 |
264774_at |
AT1G22890
|
unknown |
-0.599 |
0.444 |
| 75 |
267459_at |
AT2G33850
|
unknown |
-0.579 |
0.171 |
| 76 |
264014_at |
AT2G21210
|
SAUR6, SMALL AUXIN UPREGULATED RNA 6 |
-0.535 |
-0.217 |
| 77 |
256096_at |
AT1G13650
|
unknown |
-0.531 |
-0.608 |
| 78 |
259786_at |
AT1G29660
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.524 |
-0.414 |
| 79 |
246700_at |
AT5G28030
|
DES1, L-cysteine desulfhydrase 1 |
-0.500 |
0.275 |
| 80 |
261979_at |
AT1G37130
|
ATNR2, ARABIDOPSIS NITRATE REDUCTASE 2, B29, CHL3, CHLORATE RESISTANT 3, NIA2, nitrate reductase 2, NIA2-1, NR, NITRATE REDUCTASE, NR2, NITRATE REDUCTASE 2 |
-0.493 |
-0.077 |
| 81 |
253048_at |
AT4G37560
|
[Acetamidase/Formamidase family protein] |
-0.484 |
-0.288 |
| 82 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
-0.471 |
-0.650 |
| 83 |
246596_at |
AT5G14740
|
BETA CA2, BETA CARBONIC ANHYDRASE 2, CA18, CARBONIC ANHYDRASE 18, CA2, carbonic anhydrase 2 |
-0.457 |
-0.120 |
| 84 |
255626_at |
AT4G00780
|
[TRAF-like family protein] |
-0.437 |
0.012 |
| 85 |
259681_at |
AT1G77760
|
GNR1, NIA1, nitrate reductase 1, NR1, NITRATE REDUCTASE 1 |
-0.417 |
0.273 |
| 86 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
-0.414 |
-0.733 |
| 87 |
260774_at |
AT1G78290
|
SNRK2-8, SNF1-RELATED PROTEIN KINASE 2-8, SNRK2.8, SNF1-RELATED PROTEIN KINASE 2.8, SRK2C, SNF1-RELATED PROTEIN KINASE 2C |
-0.410 |
-0.325 |
| 88 |
256168_at |
AT1G51805
|
[Leucine-rich repeat protein kinase family protein] |
-0.405 |
0.030 |
| 89 |
267169_at |
AT2G37540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.404 |
-0.076 |
| 90 |
253806_at |
AT4G28270
|
ATRMA2, RMA2, RING membrane-anchor 2 |
-0.387 |
-0.500 |
| 91 |
253052_at |
AT4G37310
|
CYP81H1, cytochrome P450, family 81, subfamily H, polypeptide 1 |
-0.378 |
-0.044 |
| 92 |
262826_at |
AT1G13080
|
CYP71B2, cytochrome P450, family 71, subfamily B, polypeptide 2 |
-0.373 |
-0.343 |
| 93 |
264899_at |
AT1G23130
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.360 |
-0.022 |
| 94 |
249515_at |
AT5G38530
|
TSBtype2, tryptophan synthase beta type 2 |
-0.359 |
-0.052 |
| 95 |
253305_at |
AT4G33666
|
unknown |
-0.356 |
-0.156 |
| 96 |
255582_at |
AT4G01500
|
NGA4, NGATHA4 |
-0.352 |
-0.032 |
| 97 |
256891_at |
AT3G19030
|
unknown |
-0.352 |
0.190 |
| 98 |
263668_at |
AT1G04350
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.347 |
-0.156 |
| 99 |
255579_at |
AT4G01460
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.346 |
-0.423 |
| 100 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
-0.345 |
-0.780 |
| 101 |
253814_at |
AT4G28290
|
unknown |
-0.339 |
0.149 |
| 102 |
261568_at |
AT1G01030
|
NGA3, NGATHA3 |
-0.336 |
0.325 |
| 103 |
264195_at |
AT1G22690
|
[Gibberellin-regulated family protein] |
-0.335 |
-0.254 |
| 104 |
249010_at |
AT5G44580
|
unknown |
-0.329 |
-0.018 |
| 105 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
-0.321 |
1.005 |
| 106 |
259996_at |
AT1G67910
|
unknown |
-0.318 |
-0.150 |
| 107 |
248756_at |
AT5G47560
|
ATSDAT, ATTDT, TONOPLAST DICARBOXYLATE TRANSPORTER, TDT, tonoplast dicarboxylate transporter |
-0.318 |
-0.101 |
| 108 |
265698_at |
AT2G32160
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.316 |
-0.049 |
| 109 |
257017_at |
AT3G19620
|
[Glycosyl hydrolase family protein] |
-0.316 |
-0.201 |
| 110 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
-0.314 |
-0.373 |
| 111 |
246998_at |
AT5G67370
|
CGLD27, CONSERVED IN THE GREEN LINEAGE AND DIATOMS 27 |
-0.314 |
-0.308 |
| 112 |
267063_at |
AT2G41120
|
unknown |
-0.305 |
-0.156 |
| 113 |
260357_at |
AT1G69260
|
AFP1, ABI five binding protein |
-0.297 |
0.581 |
| 114 |
265475_at |
AT2G15620
|
ATHNIR, ARABIDOPSIS THALIANA NITRITE REDUCTASE, NIR, NITRITE REDUCTASE, NIR1, nitrite reductase 1 |
-0.297 |
-0.080 |
| 115 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
-0.296 |
-0.526 |
| 116 |
246195_at |
AT4G36410
|
UBC17, ubiquitin-conjugating enzyme 17 |
-0.295 |
-0.131 |
| 117 |
258038_at |
AT3G21260
|
GLTP3, GLYCOLIPID TRANSFER PROTEIN 3 |
-0.293 |
-0.235 |
| 118 |
246996_at |
AT5G67420
|
ASL39, ASYMMETRIC LEAVES2-LIKE 39, LBD37, LOB domain-containing protein 37 |
-0.291 |
-0.019 |
| 119 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
-0.290 |
-0.385 |
| 120 |
260007_at |
AT1G67870
|
[glycine-rich protein] |
-0.289 |
-0.048 |
| 121 |
254564_at |
AT4G19170
|
CCD4, carotenoid cleavage dioxygenase 4, NCED4, nine-cis-epoxycarotenoid dioxygenase 4 |
-0.286 |
-0.114 |
| 122 |
251218_at |
AT3G62410
|
CP12, CP12 DOMAIN-CONTAINING PROTEIN 1, CP12-2, CP12 domain-containing protein 2 |
-0.283 |
-0.168 |
| 123 |
261410_at |
AT1G07610
|
MT1C, metallothionein 1C |
-0.281 |
0.232 |
| 124 |
265939_at |
AT2G19650
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.280 |
-0.316 |
| 125 |
259927_at |
AT1G75100
|
JAC1, J-domain protein required for chloroplast accumulation response 1 |
-0.280 |
-0.293 |
| 126 |
261068_at |
AT1G07450
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.277 |
-0.116 |
| 127 |
267262_at |
AT2G22990
|
SCPL8, SERINE CARBOXYPEPTIDASE-LIKE 8, SNG1, sinapoylglucose 1 |
-0.277 |
0.009 |
| 128 |
250915_at |
AT5G03790
|
ATHB51, HB51, homeobox 51, LMI1, LATE MERISTEM IDENTITY1 |
-0.277 |
-0.036 |
| 129 |
251342_at |
AT3G60690
|
SAUR59, SMALL AUXIN UPREGULATED RNA 59 |
-0.276 |
0.183 |
| 130 |
249327_at |
AT5G40890
|
ATCLC-A, chloride channel A, ATCLCA, CLC-A, chloride channel A, CLC-A, CHLORIDE CHANNEL-A, CLCA, CHLORIDE CHANNEL A |
-0.274 |
-0.002 |
| 131 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
-0.274 |
0.265 |
| 132 |
246241_at |
AT4G37050
|
AtPLAIVC, phospholipase A IVC, PLA V, PLAIII{beta}, patatin-related phospholipase III beta, PLP4, PATATIN-like protein 4 |
-0.272 |
-0.094 |
| 133 |
258647_at |
AT3G07870
|
[F-box and associated interaction domains-containing protein] |
-0.271 |
-0.126 |
| 134 |
247921_at |
AT5G57660
|
ATCOL5, BBX6, B-box domain protein 6, COL5, CONSTANS-like 5 |
-0.271 |
0.184 |
| 135 |
260266_at |
AT1G68520
|
BBX14, B-box domain protein 14 |
-0.270 |
-0.459 |
| 136 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
-0.266 |
-0.037 |
| 137 |
266265_at |
AT2G29340
|
[NAD-dependent epimerase/dehydratase family protein] |
-0.263 |
0.185 |