|
probeID |
AGICode |
Annotation |
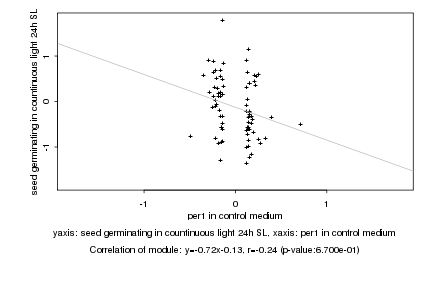
Log2 signal ratio
per1 in control medium |
Log2 signal ratio
seed germinating in countinuous light 24h SL |
| 1 |
250135_at |
AT5G15360
|
unknown |
0.708 |
-0.502 |
| 2 |
247615_at |
AT5G60250
|
[zinc finger (C3HC4-type RING finger) family protein] |
0.387 |
-0.332 |
| 3 |
260522_x_at |
AT2G41730
|
unknown |
0.328 |
-0.810 |
| 4 |
262325_at |
AT1G64160
|
AtDIR5, DIR5, dirigent protein 5 |
0.266 |
-0.915 |
| 5 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.247 |
0.605 |
| 6 |
267573_at |
AT2G30670
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.243 |
-0.831 |
| 7 |
252851_at |
AT4G40080
|
[ENTH/ANTH/VHS superfamily protein] |
0.228 |
0.562 |
| 8 |
261192_at |
AT1G32870
|
ANAC013, Arabidopsis NAC domain containing protein 13, ANAC13, NAC domain protein 13, NAC13, NAC domain protein 13 |
0.212 |
0.374 |
| 9 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
0.202 |
0.461 |
| 10 |
248727_at |
AT5G47990
|
CYP705A5, cytochrome P450, family 705, subfamily A, polypeptide 5, THAD, THALIAN-DIOL DESATURASE, THAD1, THALIAN-DIOL DESATURASE |
0.198 |
0.586 |
| 11 |
259832_at |
AT1G69580
|
[Homeodomain-like superfamily protein] |
0.191 |
-0.679 |
| 12 |
246386_at |
AT1G77410
|
BGAL16, beta-galactosidase 16 |
0.181 |
-0.383 |
| 13 |
246389_at |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
0.171 |
-0.320 |
| 14 |
256780_at |
AT3G13640
|
ABCE1, ATP-binding cassette E1, ATRLI1, RNAse l inhibitor protein 1, RLI1, RNAse l inhibitor protein 1 |
0.166 |
-0.477 |
| 15 |
245258_at |
AT4G15340
|
04C11, ATPEN1, pentacyclic triterpene synthase 1, PEN1, pentacyclic triterpene synthase 1 |
0.165 |
-1.145 |
| 16 |
256531_at |
AT1G33320
|
[Pyridoxal phosphate (PLP)-dependent transferases superfamily protein] |
0.153 |
-1.218 |
| 17 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
0.152 |
0.416 |
| 18 |
247170_at |
AT5G65530
|
[Protein kinase superfamily protein] |
0.151 |
0.415 |
| 19 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
0.148 |
-0.221 |
| 20 |
247867_at |
AT5G57630
|
CIPK21, CBL-interacting protein kinase 21, SnRK3.4, SNF1-RELATED PROTEIN KINASE 3.4 |
0.147 |
-0.597 |
| 21 |
246999_at |
AT5G67440
|
MEL2, MAB4/ENP/NPY1-LIKE 2, NPY3, NAKED PINS IN YUC MUTANTS 3 |
0.146 |
-0.204 |
| 22 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
0.144 |
-0.321 |
| 23 |
247345_at |
AT5G63760
|
ARI15, ARIADNE 15, ATARI15, ARABIDOPSIS ARIADNE 15 |
0.144 |
-0.268 |
| 24 |
250920_at |
AT5G03390
|
unknown |
0.142 |
-0.447 |
| 25 |
254728_at |
AT4G13690
|
unknown |
0.140 |
-0.568 |
| 26 |
264085_at |
AT2G31250
|
HEMA3 |
0.140 |
-0.848 |
| 27 |
247682_at |
AT5G59650
|
[Leucine-rich repeat protein kinase family protein] |
0.139 |
-0.970 |
| 28 |
249972_at |
AT5G19040
|
ATIPT5, Arabidopsis thaliana ISOPENTENYLTRANSFERASE 5, IPT5, isopentenyltransferase 5 |
0.137 |
-0.601 |
| 29 |
262795_at |
AT1G13130
|
[Cellulase (glycosyl hydrolase family 5) protein] |
0.133 |
-0.340 |
| 30 |
250941_at |
AT5G03320
|
[Protein kinase superfamily protein] |
0.133 |
1.153 |
| 31 |
251282_at |
AT3G61630
|
CRF6, cytokinin response factor 6 |
0.130 |
-0.720 |
| 32 |
260303_at |
AT1G70520
|
ASG6, ALTERED SEED GERMINATION 6, CRK2, cysteine-rich RLK (RECEPTOR-like protein kinase) 2 |
0.129 |
0.640 |
| 33 |
248814_at |
AT5G46910
|
[Transcription factor jumonji (jmj) family protein / zinc finger (C5HC2 type) family protein] |
0.123 |
0.049 |
| 34 |
247393_at |
AT5G63130
|
[Octicosapeptide/Phox/Bem1p family protein] |
0.122 |
-0.551 |
| 35 |
256574_at |
AT3G14780
|
unknown |
0.121 |
-1.356 |
| 36 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
0.121 |
-0.206 |
| 37 |
261894_at |
AT1G80900
|
ATMGT1, MAGNESIUM TRANSPORTER 1, MGT1, magnesium transporter 1, MRS2-10 |
0.121 |
-0.075 |
| 38 |
264040_at |
AT2G03730
|
ACR5, ACT domain repeat 5 |
0.119 |
0.915 |
| 39 |
245619_at |
AT4G13990
|
[Exostosin family protein] |
0.117 |
0.321 |
| 40 |
254404_at |
AT4G21340
|
B70 |
0.116 |
-0.621 |
| 41 |
263658_at |
AT1G04490
|
unknown |
0.115 |
-0.993 |
| 42 |
260150_at |
AT1G52820
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.497 |
-0.764 |
| 43 |
264672_at |
AT1G09750
|
[Eukaryotic aspartyl protease family protein] |
-0.355 |
0.588 |
| 44 |
247320_at |
AT5G64040
|
PSAN |
-0.295 |
0.922 |
| 45 |
264023_at |
AT2G21195
|
unknown |
-0.286 |
0.210 |
| 46 |
249557_at |
AT5G38280
|
PR5K, PR5-like receptor kinase |
-0.260 |
-0.121 |
| 47 |
250257_at |
AT5G13770
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
-0.250 |
0.122 |
| 48 |
249524_at |
AT5G38520
|
[alpha/beta-Hydrolases superfamily protein] |
-0.248 |
0.657 |
| 49 |
246330_at |
AT3G43600
|
AAO2, aldehyde oxidase 2, AO3, aldehyde oxidase 3, AOgamma, Aldehyde oxidase gamma, atAO-2, Arabidopsis thaliana aldehyde oxidase 2, AtAO3, Arabidopsis thaliana aldehyde oxidase 3 |
-0.245 |
0.893 |
| 50 |
257054_at |
AT3G15353
|
ATMT3, MT3, metallothionein 3 |
-0.236 |
0.325 |
| 51 |
264545_at |
AT1G55670
|
PSAG, photosystem I subunit G |
-0.228 |
0.684 |
| 52 |
251617_at |
AT3G58000
|
[VQ motif-containing protein] |
-0.221 |
-0.795 |
| 53 |
245774_at |
AT1G30210
|
ATTCP24, TCP24, TEOSINTE BRANCHED 1, cycloidea, and PCF family 24 |
-0.219 |
-0.108 |
| 54 |
254836_at |
AT4G12330
|
CYP706A7, cytochrome P450, family 706, subfamily A, polypeptide 7 |
-0.219 |
0.034 |
| 55 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
-0.216 |
0.510 |
| 56 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
-0.215 |
-0.049 |
| 57 |
245106_at |
AT2G41650
|
unknown |
-0.206 |
0.294 |
| 58 |
252448_at |
AT3G47050
|
[Glycosyl hydrolase family protein] |
-0.194 |
-0.902 |
| 59 |
259737_at |
AT1G64400
|
LACS3, long-chain acyl-CoA synthetase 3 |
-0.187 |
0.131 |
| 60 |
259996_at |
AT1G67910
|
unknown |
-0.186 |
0.185 |
| 61 |
264282_at |
AT1G61840
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.177 |
-0.195 |
| 62 |
246906_at |
AT5G25475
|
[AP2/B3-like transcriptional factor family protein] |
-0.173 |
0.200 |
| 63 |
261530_at |
AT1G63460
|
ATGPX8, GPX8, glutathione peroxidase 8 |
-0.173 |
0.114 |
| 64 |
264301_at |
AT1G78780
|
[pathogenesis-related family protein] |
-0.173 |
-0.313 |
| 65 |
251031_at |
AT5G02120
|
OHP, one helix protein, PDE335, PIGMENT DEFECTIVE 335 |
-0.172 |
0.700 |
| 66 |
254286_at |
AT4G22950
|
AGL19, AGAMOUS-like 19, GL19 |
-0.168 |
-1.277 |
| 67 |
262970_at |
AT1G75690
|
LQY1, LOW QUANTUM YIELD OF PHOTOSYSTEM II 1 |
-0.166 |
0.550 |
| 68 |
255527_at |
AT4G02360
|
unknown |
-0.160 |
-0.569 |
| 69 |
266353_at |
AT2G01520
|
MLP328, MLP-like protein 328, ZCE1, (Zusammen-CA)-enhanced 1 |
-0.160 |
-0.900 |
| 70 |
245300_at |
AT4G16350
|
CBL6, calcineurin B-like protein 6, SCABP2, SOS3-LIKE CALCIUM BINDING PROTEIN 2 |
-0.157 |
-0.898 |
| 71 |
249033_at |
AT5G44920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.154 |
-0.570 |
| 72 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
-0.148 |
-0.468 |
| 73 |
250180_at |
AT5G14450
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.146 |
1.802 |
| 74 |
246235_at |
AT4G36830
|
HOS3-1 |
-0.145 |
0.172 |
| 75 |
245730_at |
AT1G73470
|
unknown |
-0.144 |
0.163 |
| 76 |
263918_at |
AT2G36590
|
ATPROT3, PROLINE TRANSPORTER 3, ProT3, proline transporter 3 |
-0.144 |
0.491 |
| 77 |
256100_at |
AT1G13750
|
[Purple acid phosphatases superfamily protein] |
-0.144 |
-0.310 |
| 78 |
258738_at |
AT3G05750
|
TRM6, TON1 Recruiting Motif 6 |
-0.144 |
-0.873 |
| 79 |
248770_at |
AT5G47740
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.142 |
-0.612 |
| 80 |
246891_at |
AT5G25490
|
[Ran BP2/NZF zinc finger-like superfamily protein] |
-0.141 |
0.842 |
| 81 |
253566_at |
AT4G31210
|
[DNA topoisomerase, type IA, core] |
-0.140 |
0.352 |