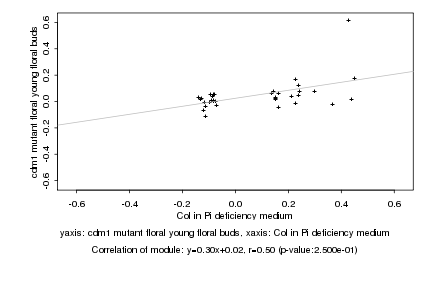
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Col in Pi deficiency medium |
Log2 signal ratio
cdm1 mutant floral young floral buds |
| 1 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.449 |
0.178 |
| 2 |
252988_at |
AT4G38410
|
[Dehydrin family protein] |
0.435 |
0.021 |
| 3 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.426 |
0.622 |
| 4 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
0.366 |
-0.022 |
| 5 |
256319_at |
AT1G35910
|
TPPD, trehalose-6-phosphate phosphatase D |
0.298 |
0.082 |
| 6 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.239 |
0.079 |
| 7 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
0.237 |
0.124 |
| 8 |
260401_at |
AT1G69840
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
0.235 |
0.049 |
| 9 |
251884_at |
AT3G54150
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.225 |
-0.013 |
| 10 |
246001_at |
AT5G20790
|
unknown |
0.224 |
0.169 |
| 11 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
0.210 |
0.044 |
| 12 |
267075_at |
AT2G41070
|
ATBZIP12, DPBF4, EEL, ENHANCED EM LEVEL |
0.161 |
-0.041 |
| 13 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.159 |
0.066 |
| 14 |
251885_at |
AT3G54050
|
HCEF1, high cyclic electron flow 1 |
0.148 |
0.019 |
| 15 |
247205_at |
AT5G64890
|
PROPEP2, elicitor peptide 2 precursor |
0.148 |
0.024 |
| 16 |
247047_at |
AT5G66650
|
unknown |
0.148 |
0.032 |
| 17 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
0.141 |
0.079 |
| 18 |
261203_at |
AT1G12845
|
unknown |
0.136 |
0.068 |
| 19 |
245577_at |
AT4G14780
|
[Protein kinase superfamily protein] |
-0.140 |
0.031 |
| 20 |
247410_at |
AT5G62990
|
emb1692, embryo defective 1692 |
-0.133 |
0.016 |
| 21 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.131 |
0.027 |
| 22 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
-0.124 |
-0.063 |
| 23 |
247586_at |
AT5G60660
|
PIP2;4, plasma membrane intrinsic protein 2;4, PIP2F |
-0.120 |
-0.002 |
| 24 |
266990_at |
AT2G39190
|
ATATH8 |
-0.116 |
-0.033 |
| 25 |
245737_at |
AT1G44160
|
[HSP40/DnaJ peptide-binding protein] |
-0.114 |
-0.108 |
| 26 |
259030_at |
AT3G09310
|
unknown |
-0.098 |
-0.004 |
| 27 |
258562_at |
AT3G05980
|
unknown |
-0.096 |
0.060 |
| 28 |
250257_at |
AT5G13770
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
-0.091 |
0.015 |
| 29 |
252599_at |
AT3G45390
|
LecRK-I.2, L-type lectin receptor kinase I.2 |
-0.089 |
0.041 |
| 30 |
261338_at |
AT1G44920
|
unknown |
-0.084 |
0.001 |
| 31 |
267586_at |
AT2G41950
|
unknown |
-0.084 |
0.055 |
| 32 |
249729_at |
AT5G24410
|
PGL4, 6-phosphogluconolactonase 4 |
-0.083 |
0.013 |
| 33 |
255484_at |
AT4G02540
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.080 |
0.060 |
| 34 |
249762_at |
AT5G24000
|
unknown |
-0.078 |
0.002 |
| 35 |
257236_at |
AT3G15095
|
HCF243, high chlorophyll fluorescence 243 |
-0.073 |
-0.026 |