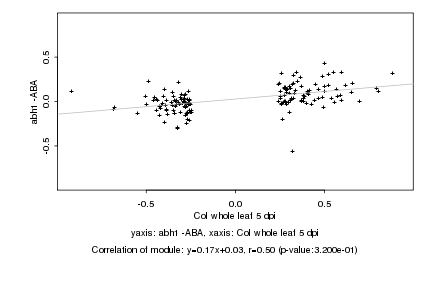
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Col whole leaf 5 dpi |
Log2 signal ratio
abh1 -ABA |
| 1 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.876 |
0.316 |
| 2 |
259559_at |
AT1G21240
|
WAK3, wall associated kinase 3 |
0.799 |
0.122 |
| 3 |
246368_at |
AT1G51890
|
[Leucine-rich repeat protein kinase family protein] |
0.786 |
0.151 |
| 4 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
0.695 |
0.008 |
| 5 |
245076_at |
AT2G23170
|
GH3.3 |
0.651 |
0.204 |
| 6 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
0.646 |
0.110 |
| 7 |
255340_at |
AT4G04490
|
CRK36, cysteine-rich RLK (RECEPTOR-like protein kinase) 36 |
0.614 |
0.181 |
| 8 |
257100_at |
AT3G25010
|
AtRLP41, receptor like protein 41, RLP41, receptor like protein 41 |
0.594 |
0.330 |
| 9 |
253687_at |
AT4G29520
|
[LOCATED IN: endoplasmic reticulum, plasma membrane] |
0.590 |
0.022 |
| 10 |
261394_at |
AT1G79680
|
ATWAKL10, WAKL10, WALL ASSOCIATED KINASE (WAK)-LIKE 10 |
0.587 |
0.077 |
| 11 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
0.571 |
0.062 |
| 12 |
258941_at |
AT3G09940
|
ATMDAR3, ARABIDOPSIS THALIANA MONODEHYDROASCORBATE REDUCTASE 3, MDAR2, MONODEHYDROASCORBATE REDUCTASE 2, MDAR3, MONODEHYDROASCORBATE REDUCTASE 3, MDHAR, monodehydroascorbate reductase |
0.564 |
0.144 |
| 13 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
0.554 |
-0.000 |
| 14 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.546 |
0.329 |
| 15 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.538 |
0.042 |
| 16 |
254571_at |
AT4G19370
|
unknown |
0.518 |
0.185 |
| 17 |
247314_at |
AT5G64000
|
ATSAL2, SAL2 |
0.518 |
0.306 |
| 18 |
255341_at |
AT4G04500
|
CRK37, cysteine-rich RLK (RECEPTOR-like protein kinase) 37 |
0.497 |
0.436 |
| 19 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.496 |
0.123 |
| 20 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.495 |
0.178 |
| 21 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
0.492 |
-0.057 |
| 22 |
261216_at |
AT1G33030
|
[O-methyltransferase family protein] |
0.486 |
0.046 |
| 23 |
259489_at |
AT1G15790
|
unknown |
0.485 |
0.285 |
| 24 |
261167_at |
AT1G04980
|
ATPDI10, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 10, ATPDIL2-2, PDI-like 2-2, PDI10, PROTEIN DISULFIDE ISOMERASE, PDIL2-2, PDI-like 2-2 |
0.465 |
0.043 |
| 25 |
263847_at |
AT2G36970
|
[UDP-Glycosyltransferase superfamily protein] |
0.464 |
0.145 |
| 26 |
267300_at |
AT2G30140
|
UGT87A2, UDP-glucosyl transferase 87A2 |
0.446 |
0.200 |
| 27 |
247435_at |
AT5G62480
|
ATGSTU9, glutathione S-transferase tau 9, GST14, GLUTATHIONE S-TRANSFERASE 14, GST14B, GLUTATHIONE S-TRANSFERASE 14B, GSTU9, glutathione S-transferase tau 9 |
0.441 |
0.022 |
| 28 |
255912_at |
AT1G66960
|
LUP5 |
0.423 |
-0.029 |
| 29 |
260239_at |
AT1G74360
|
[Leucine-rich repeat protein kinase family protein] |
0.412 |
0.126 |
| 30 |
251884_at |
AT3G54150
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.409 |
0.104 |
| 31 |
266782_at |
AT2G29120
|
ATGLR2.7, glutamate receptor 2.7, GLR2.7, GLUTAMATE RECEPTOR 2.7, GLR2.7, glutamate receptor 2.7 |
0.406 |
0.086 |
| 32 |
253181_at |
AT4G35180
|
LHT7, LYS/HIS transporter 7 |
0.403 |
0.100 |
| 33 |
267546_at |
AT2G32680
|
AtRLP23, receptor like protein 23, RLP23, receptor like protein 23 |
0.400 |
0.121 |
| 34 |
267413_at |
AT2G34960
|
CAT5, cationic amino acid transporter 5 |
0.396 |
-0.012 |
| 35 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
0.382 |
0.061 |
| 36 |
257154_at |
AT3G27210
|
unknown |
0.378 |
0.077 |
| 37 |
256299_at |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
0.376 |
0.002 |
| 38 |
248060_at |
AT5G55560
|
[Protein kinase superfamily protein] |
0.376 |
0.036 |
| 39 |
267436_at |
AT2G19190
|
FRK1, FLG22-induced receptor-like kinase 1 |
0.370 |
0.019 |
| 40 |
258385_at |
AT3G15510
|
ANAC056, Arabidopsis NAC domain containing protein 56, ATNAC2, NAC domain containing protein 2, NAC2, NAC domain containing protein 2, NARS1, NAC-REGULATED SEED MORPHOLOGY 1 |
0.369 |
0.002 |
| 41 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
0.366 |
0.170 |
| 42 |
265161_at |
AT1G30900
|
BP80-3;3, binding protein of 80 kDa 3;3, VSR3;3, VACUOLAR SORTING RECEPTOR 3;3, VSR6, VACUOLAR SORTING RECEPTOR 6 |
0.360 |
0.275 |
| 43 |
263852_at |
AT2G04450
|
ATNUDT6, nudix hydrolase homolog 6, ATNUDX6, Arabidopsis thaliana nucleoside diphosphate linked to some moiety X 6, NUDT6, nudix hydrolase homolog 6, NUDX6, nucleoside diphosphates linked to some moiety X 6 |
0.347 |
0.232 |
| 44 |
252060_at |
AT3G52430
|
ATPAD4, ARABIDOPSIS PHYTOALEXIN DEFICIENT 4, PAD4, PHYTOALEXIN DEFICIENT 4 |
0.339 |
0.330 |
| 45 |
246988_at |
AT5G67340
|
[ARM repeat superfamily protein] |
0.335 |
0.129 |
| 46 |
252825_at |
AT4G39890
|
AtRABH1c, RAB GTPase homolog H1C, RABH1c, RAB GTPase homolog H1C |
0.331 |
0.098 |
| 47 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
0.325 |
0.298 |
| 48 |
248333_at |
AT5G52390
|
[PAR1 protein] |
0.322 |
0.206 |
| 49 |
265189_at |
AT1G23840
|
unknown |
0.321 |
0.045 |
| 50 |
257625_at |
AT3G26230
|
CYP71B24, cytochrome P450, family 71, subfamily B, polypeptide 24 |
0.317 |
-0.559 |
| 51 |
251673_at |
AT3G57240
|
AtBG3, BG3, beta-1,3-glucanase 3 |
0.316 |
0.199 |
| 52 |
261023_at |
AT1G12200
|
FMO, flavin monooxygenase |
0.311 |
0.042 |
| 53 |
265356_at |
AT2G16595
|
[Translocon-associated protein (TRAP), alpha subunit] |
0.309 |
0.031 |
| 54 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.307 |
0.156 |
| 55 |
255794_at |
AT2G33480
|
ANAC041, NAC domain containing protein 41, NAC041, NAC domain containing protein 41 |
0.304 |
0.014 |
| 56 |
250267_at |
AT5G12930
|
unknown |
0.303 |
0.174 |
| 57 |
266017_at |
AT2G18690
|
unknown |
0.303 |
0.098 |
| 58 |
256697_at |
AT3G20660
|
AtOCT4, organic cation/carnitine transporter4, OCT4, organic cation/carnitine transporter4 |
0.303 |
-0.119 |
| 59 |
249309_at |
AT5G41410
|
BEL1, BELL 1 |
0.296 |
-0.005 |
| 60 |
249422_at |
AT5G39760
|
AtHB23, homeobox protein 23, HB23, homeobox protein 23, ZHD10, ZINC FINGER HOMEODOMAIN 10 |
0.295 |
-0.003 |
| 61 |
254500_at |
AT4G20110
|
BP80-3;1, binding protein of 80 kDa 3;1, VSR3;1, VACUOLAR SORTING RECEPTOR 3;1, VSR7, VACUOLAR SORTING RECEPTOR 7 |
0.288 |
0.145 |
| 62 |
259852_at |
AT1G72280
|
AERO1, endoplasmic reticulum oxidoreductins 1, ERO1, endoplasmic reticulum oxidoreductins 1 |
0.285 |
0.104 |
| 63 |
250400_at |
AT5G10740
|
[Protein phosphatase 2C family protein] |
0.285 |
0.137 |
| 64 |
261456_at |
AT1G21050
|
unknown |
0.282 |
-0.030 |
| 65 |
266167_at |
AT2G38860
|
DJ-1e, DJ1E, YLS5 |
0.279 |
0.004 |
| 66 |
262378_at |
AT1G72830
|
ATHAP2C, HAP2C, NF-YA3, nuclear factor Y, subunit A3 |
0.277 |
0.140 |
| 67 |
257535_at |
AT3G09490
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.275 |
0.164 |
| 68 |
251342_at |
AT3G60690
|
SAUR59, SMALL AUXIN UPREGULATED RNA 59 |
0.275 |
-0.010 |
| 69 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
0.274 |
0.156 |
| 70 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
0.273 |
0.011 |
| 71 |
259598_at |
AT1G27980
|
ATDPL1, DPL1, dihydrosphingosine phosphate lyase |
0.273 |
0.074 |
| 72 |
265620_at |
AT2G27310
|
[F-box family protein] |
0.271 |
0.158 |
| 73 |
253519_at |
AT4G31240
|
AtNRX2, NRX2, nucleoredoxin 2 |
0.265 |
-0.009 |
| 74 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.260 |
-0.200 |
| 75 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
0.256 |
0.317 |
| 76 |
246098_at |
AT5G20400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.254 |
-0.027 |
| 77 |
245431_at |
AT4G17080
|
[Histone H3 K4-specific methyltransferase SET7/9 family protein] |
0.254 |
-0.013 |
| 78 |
263605_at |
AT2G16485
|
NERD, Needed for RDR2-independent DNA methylation |
0.252 |
0.042 |
| 79 |
251282_at |
AT3G61630
|
CRF6, cytokinin response factor 6 |
0.251 |
0.059 |
| 80 |
263183_at |
AT1G05570
|
ATGSL06, ATGSL6, CALS1, callose synthase 1, GSL06, GSL6, GLUCAN SYNTHASE-LIKE 6 |
0.251 |
0.117 |
| 81 |
245293_at |
AT4G16660
|
[heat shock protein 70 (Hsp 70) family protein] |
0.244 |
0.206 |
| 82 |
260201_at |
AT1G67600
|
[Acid phosphatase/vanadium-dependent haloperoxidase-related protein] |
0.239 |
0.006 |
| 83 |
262177_at |
AT1G74710
|
ATICS1, ARABIDOPSIS ISOCHORISMATE SYNTHASE 1, EDS16, ENHANCED DISEASE SUSCEPTIBILITY TO ERYSIPHE ORONTII 16, ICS1, ISOCHORISMATE SYNTHASE 1, SID2, SALICYLIC ACID INDUCTION DEFICIENT 2 |
0.239 |
0.192 |
| 84 |
246004_at |
AT5G20630
|
ATGER3, ARABIDOPSIS THALIANA GERMIN 3, GER3, germin 3, GLP3, GERMIN-LIKE PROTEIN 3, GLP3A, GLP3B |
-0.923 |
0.113 |
| 85 |
260982_at |
AT1G53520
|
AtFAP3, FAP3, fatty-acid-binding protein 3 |
-0.686 |
-0.079 |
| 86 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
-0.682 |
-0.059 |
| 87 |
245980_at |
AT5G13140
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.554 |
-0.130 |
| 88 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
-0.507 |
0.067 |
| 89 |
252997_at |
AT4G38400
|
ATEXLA2, expansin-like A2, ATEXPL2, ATHEXP BETA 2.2, EXLA2, expansin-like A2, EXPL2, EXPANSIN L2 |
-0.499 |
-0.025 |
| 90 |
266545_at |
AT2G35290
|
SAUR79, SMALL AUXIN UPREGULATED RNA 79 |
-0.491 |
0.232 |
| 91 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
-0.461 |
0.017 |
| 92 |
263480_at |
AT2G04032
|
ZIP7, zinc transporter 7 precursor |
-0.459 |
0.048 |
| 93 |
245196_at |
AT1G67750
|
[Pectate lyase family protein] |
-0.447 |
-0.092 |
| 94 |
260028_at |
AT1G29980
|
unknown |
-0.445 |
0.029 |
| 95 |
262830_at |
AT1G14700
|
ATPAP3, PAP3, purple acid phosphatase 3 |
-0.441 |
0.018 |
| 96 |
255549_at |
AT4G01950
|
ATGPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3, GPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3 |
-0.429 |
-0.045 |
| 97 |
266715_at |
AT2G46780
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.428 |
-0.150 |
| 98 |
249817_at |
AT5G23820
|
ML3, MD2-related lipid recognition 3 |
-0.424 |
-0.077 |
| 99 |
253191_at |
AT4G35350
|
XCP1, xylem cysteine peptidase 1 |
-0.419 |
-0.035 |
| 100 |
266447_at |
AT2G43290
|
MSS3, multicopy suppressors of snf4 deficiency in yeast 3 |
-0.411 |
-0.014 |
| 101 |
246200_at |
AT4G37240
|
unknown |
-0.404 |
0.063 |
| 102 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
-0.402 |
-0.225 |
| 103 |
246993_at |
AT5G67450
|
AZF1, zinc-finger protein 1, ZF1, zinc-finger protein 1 |
-0.401 |
0.137 |
| 104 |
263628_at |
AT2G04780
|
FLA7, FASCICLIN-like arabinoogalactan 7 |
-0.396 |
-0.037 |
| 105 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
-0.392 |
-0.095 |
| 106 |
260547_at |
AT2G43550
|
[Scorpion toxin-like knottin superfamily protein] |
-0.388 |
-0.087 |
| 107 |
262354_at |
AT1G64200
|
VHA-E3, vacuolar H+-ATPase subunit E isoform 3 |
-0.388 |
0.021 |
| 108 |
253643_at |
AT4G29780
|
unknown |
-0.381 |
-0.144 |
| 109 |
249583_at |
AT5G37770
|
CML24, CALMODULIN-LIKE 24, TCH2, TOUCH 2 |
-0.361 |
-0.003 |
| 110 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
-0.356 |
-0.042 |
| 111 |
251955_at |
AT3G53680
|
[Acyl-CoA N-acyltransferase with RING/FYVE/PHD-type zinc finger domain] |
-0.353 |
0.106 |
| 112 |
252377_at |
AT3G47960
|
AtNPF2.10, GTR1, GLUCOSINOLATE TRANSPORTER-1, NPF2.10, NRT1/ PTR family 2.10 |
-0.351 |
0.059 |
| 113 |
250006_at |
AT5G18660
|
PCB2, PALE-GREEN AND CHLOROPHYLL B REDUCED 2 |
-0.350 |
-0.096 |
| 114 |
266418_at |
AT2G38750
|
ANNAT4, annexin 4, AtANN4 |
-0.345 |
0.015 |
| 115 |
248975_at |
AT5G45040
|
CYTC6A, cytochrome c6A |
-0.345 |
-0.124 |
| 116 |
252374_at |
AT3G48100
|
ARR5, response regulator 5, ATRR2, ARABIDOPSIS THALIANA RESPONSE REGULATOR 2, IBC6, INDUCED BY CYTOKININ 6, RR5, response regulator 5 |
-0.342 |
-0.125 |
| 117 |
252365_at |
AT3G48350
|
CEP3, cysteine endopeptidase 3 |
-0.340 |
0.002 |
| 118 |
250933_at |
AT5G03170
|
ATFLA11, ARABIDOPSIS FASCICLIN-LIKE ARABINOGALACTAN-PROTEIN 11, FLA11, FASCICLIN-like arabinogalactan-protein 11 |
-0.340 |
-0.055 |
| 119 |
263726_at |
AT2G13610
|
ABCG5, ATP-binding cassette G5 |
-0.335 |
-0.030 |
| 120 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
-0.333 |
0.022 |
| 121 |
247100_at |
AT5G66520
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.329 |
-0.294 |
| 122 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.326 |
-0.289 |
| 123 |
261175_at |
AT1G04800
|
[glycine-rich protein] |
-0.321 |
0.223 |
| 124 |
252943_at |
AT4G39330
|
ATCAD9, CAD9, cinnamyl alcohol dehydrogenase 9 |
-0.320 |
-0.003 |
| 125 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.314 |
-0.026 |
| 126 |
247524_at |
AT5G61440
|
ACHT5, atypical CYS HIS rich thioredoxin 5 |
-0.309 |
0.053 |
| 127 |
262290_at |
AT1G70985
|
[hydroxyproline-rich glycoprotein family protein] |
-0.309 |
-0.116 |
| 128 |
252143_at |
AT3G51150
|
[ATP binding microtubule motor family protein] |
-0.306 |
0.024 |
| 129 |
251846_at |
AT3G54560
|
HTA11, histone H2A 11 |
-0.303 |
0.082 |
| 130 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.300 |
-0.018 |
| 131 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
-0.299 |
-0.019 |
| 132 |
250661_at |
AT5G07030
|
[Eukaryotic aspartyl protease family protein] |
-0.296 |
0.027 |
| 133 |
259979_at |
AT1G76600
|
unknown |
-0.296 |
0.005 |
| 134 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
-0.289 |
0.038 |
| 135 |
249932_at |
AT5G22390
|
unknown |
-0.289 |
0.076 |
| 136 |
253480_at |
AT4G31840
|
AtENODL15, ENODL15, early nodulin-like protein 15 |
-0.287 |
-0.049 |
| 137 |
262414_at |
AT1G49430
|
LACS2, long-chain acyl-CoA synthetase 2, LRD2, LATERAL ROOT DEVELOPMENT 2 |
-0.287 |
0.005 |
| 138 |
264096_at |
AT1G78995
|
unknown |
-0.285 |
-0.147 |
| 139 |
261801_at |
AT1G30520
|
AAE14, acyl-activating enzyme 14 |
-0.285 |
-0.150 |
| 140 |
265118_at |
AT1G62660
|
[Glycosyl hydrolases family 32 protein] |
-0.284 |
0.086 |
| 141 |
261256_at |
AT1G05760
|
RTM1, restricted tev movement 1 |
-0.283 |
-0.021 |
| 142 |
264179_at |
AT1G02180
|
[ferredoxin-related] |
-0.282 |
-0.061 |
| 143 |
248597_at |
AT5G49160
|
DDM2, DECREASED DNA METHYLATION 2, DMT01, DNA METHYLTRANSFERASE 01, DMT1, DNA METHYLTRANSFERASE 1, MET1, methyltransferase 1, MET2, METHYLTRANSFERASE 2, METI, METHYLTRANSFERASE I |
-0.280 |
-0.007 |
| 144 |
250820_at |
AT5G05160
|
RUL1, REDUCED IN LATERAL GROWTH1 |
-0.280 |
-0.065 |
| 145 |
245743_at |
AT1G51080
|
unknown |
-0.278 |
-0.238 |
| 146 |
259014_at |
AT3G07320
|
[O-Glycosyl hydrolases family 17 protein] |
-0.277 |
-0.132 |
| 147 |
261570_at |
AT1G01120
|
KCS1, 3-ketoacyl-CoA synthase 1 |
-0.277 |
-0.046 |
| 148 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
-0.274 |
-0.119 |
| 149 |
267038_at |
AT2G38480
|
[Uncharacterised protein family (UPF0497)] |
-0.272 |
-0.132 |
| 150 |
261949_at |
AT1G64670
|
BDG1, BODYGUARD1, CED1, 9-CIS EPOXYCAROTENOID DIOXYGENASE DEFECTIVE 1 |
-0.272 |
0.032 |
| 151 |
253522_at |
AT4G31290
|
AtGGCT2;2, GGCT2;2, gamma-glutamyl cyclotransferase 2;2 |
-0.269 |
-0.194 |
| 152 |
247213_at |
AT5G64900
|
ATPEP1, ARABIDOPSIS THALIANA PEPTIDE 1, PEP1, PEPTIDE 1, PROPEP1, precursor of peptide 1 |
-0.268 |
0.124 |
| 153 |
263127_at |
AT1G78610
|
MSL6, mechanosensitive channel of small conductance-like 6 |
-0.266 |
-0.032 |
| 154 |
265732_at |
AT2G01300
|
unknown |
-0.264 |
-0.009 |
| 155 |
251395_at |
AT2G45470
|
AGP8, ARABINOGALACTAN PROTEIN 8, FLA8, FASCICLIN-like arabinogalactan protein 8 |
-0.263 |
-0.016 |
| 156 |
247977_at |
AT5G56850
|
unknown |
-0.263 |
-0.209 |
| 157 |
251899_at |
AT3G54400
|
[Eukaryotic aspartyl protease family protein] |
-0.260 |
0.028 |
| 158 |
251982_at |
AT3G53190
|
[Pectin lyase-like superfamily protein] |
-0.260 |
-0.014 |
| 159 |
249061_at |
AT5G44550
|
[Uncharacterised protein family (UPF0497)] |
-0.258 |
0.012 |
| 160 |
251714_at |
AT3G56370
|
[Leucine-rich repeat protein kinase family protein] |
-0.258 |
-0.030 |
| 161 |
248094_at |
AT5G55220
|
[trigger factor type chaperone family protein] |
-0.258 |
-0.097 |
| 162 |
264315_at |
AT1G70370
|
PG2, polygalacturonase 2 |
-0.255 |
-0.033 |
| 163 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
-0.253 |
-0.121 |
| 164 |
249278_at |
AT5G41900
|
[alpha/beta-Hydrolases superfamily protein] |
-0.248 |
-0.123 |
| 165 |
253579_at |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
-0.248 |
-0.093 |