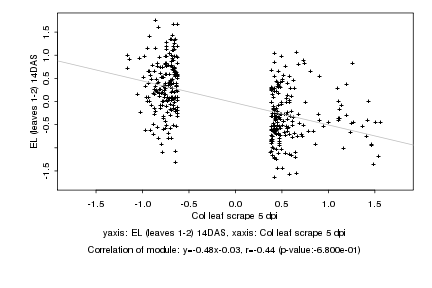
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Col leaf scrape 5 dpi |
Log2 signal ratio
EL (leaves 1-2) 14DAS |
| 1 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
1.555 |
-0.433 |
| 2 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
1.533 |
-1.186 |
| 3 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
1.502 |
-0.438 |
| 4 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
1.477 |
-1.360 |
| 5 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
1.462 |
-0.950 |
| 6 |
260978_at |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
1.456 |
-0.920 |
| 7 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
1.422 |
0.009 |
| 8 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
1.420 |
-0.396 |
| 9 |
261838_at |
AT1G16030
|
Hsp70b, heat shock protein 70B |
1.409 |
-0.755 |
| 10 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
1.358 |
-0.529 |
| 11 |
254839_at |
AT4G12400
|
Hop3, Hop3 |
1.265 |
-0.451 |
| 12 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
1.254 |
0.828 |
| 13 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
1.246 |
-0.460 |
| 14 |
257337_at |
ATMG00060
|
NAD5, NADH DEHYDROGENASE SUBUNIT 5, NAD5.3, NADH DEHYDROGENASE SUBUNIT 5.3, NAD5C, NADH dehydrogenase subunit 5C |
1.235 |
-0.648 |
| 15 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
1.193 |
0.374 |
| 16 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
1.193 |
-0.298 |
| 17 |
266590_at |
AT2G46240
|
ATBAG6, ARABIDOPSIS THALIANA BCL-2-ASSOCIATED ATHANOGENE 6, BAG6, BCL-2-associated athanogene 6 |
1.161 |
-1.000 |
| 18 |
264617_at |
AT2G17660
|
[RPM1-interacting protein 4 (RIN4) family protein] |
1.134 |
-0.068 |
| 19 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
1.117 |
-0.348 |
| 20 |
253884_at |
AT4G27670
|
HSP21, heat shock protein 21 |
1.111 |
-0.170 |
| 21 |
260025_at |
AT1G30070
|
[SGS domain-containing protein] |
1.104 |
0.001 |
| 22 |
265675_at |
AT2G32120
|
HSP70T-2, heat-shock protein 70T-2 |
1.103 |
-0.347 |
| 23 |
244902_at |
ATMG00650
|
NAD4L, NADH dehydrogenase subunit 4L |
1.099 |
-0.393 |
| 24 |
258336_at |
AT3G16050
|
A37, ATPDX1.2, ARABIDOPSIS THALIANA PYRIDOXINE BIOSYNTHESIS 1.2, PDX1.2, pyridoxine biosynthesis 1.2 |
1.088 |
0.282 |
| 25 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.992 |
-0.443 |
| 26 |
246944_at |
AT5G25450
|
[Cytochrome bd ubiquinol oxidase, 14kDa subunit] |
0.945 |
-0.531 |
| 27 |
255891_at |
AT1G17870
|
ATEGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3, EGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3 |
0.903 |
0.541 |
| 28 |
244943_at |
ATMG00070
|
NAD9, NADH dehydrogenase subunit 9 |
0.894 |
-0.395 |
| 29 |
250502_at |
AT5G09590
|
HSC70-5, HEAT SHOCK COGNATE, MTHSC70-2, mitochondrial HSO70 2 |
0.887 |
-0.270 |
| 30 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
0.858 |
-0.912 |
| 31 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
0.837 |
-0.638 |
| 32 |
260425_at |
AT1G72440
|
EDA25, embryo sac development arrest 25, SWA2, SLOW WALKER2 |
0.803 |
0.663 |
| 33 |
256347_at |
AT1G54923
|
unknown |
0.783 |
-0.630 |
| 34 |
261839_at |
AT1G16040
|
unknown |
0.753 |
-0.001 |
| 35 |
257822_at |
AT3G25230
|
ATFKBP62, FKBP62, FK506 BINDING PROTEIN 62, ROF1, rotamase FKBP 1 |
0.734 |
0.823 |
| 36 |
244906_at |
ATMG00690
|
ORF240A |
0.731 |
-0.513 |
| 37 |
256983_at |
AT3G13470
|
Cpn60beta2, chaperonin-60beta2 |
0.729 |
0.898 |
| 38 |
264035_at |
AT2G03630
|
unknown |
0.711 |
-0.753 |
| 39 |
257332_at |
ATMG01350
|
ORF145C |
0.705 |
-0.707 |
| 40 |
256392_at |
AT3G06260
|
GATL4, galacturonosyltransferase-like 4, GolS9, galactinol synthase 9 |
0.681 |
-0.466 |
| 41 |
254550_at |
AT4G19690
|
ATIRT1, ARABIDOPSIS IRON-REGULATED TRANSPORTER 1, IRT1, iron-regulated transporter 1 |
0.671 |
-0.264 |
| 42 |
250994_at |
AT5G02490
|
AtHsp70-2, Hsp70-2 |
0.668 |
0.813 |
| 43 |
255165_at |
AT4G07890
|
[gypsy-like retrotransposon family (Athila), has a 1.2e-14 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
0.664 |
-0.740 |
| 44 |
266784_at |
AT2G28960
|
[Leucine-rich repeat protein kinase family protein] |
0.656 |
-1.541 |
| 45 |
254076_at |
AT4G25340
|
ATFKBP53, FKBP53, FK506 BINDING PROTEIN 53 |
0.652 |
1.076 |
| 46 |
259552_at |
AT1G21320
|
[nucleotide binding] |
0.645 |
-1.097 |
| 47 |
248513_at |
AT5G50500
|
unknown |
0.639 |
-1.208 |
| 48 |
259635_at |
AT1G56360
|
ATPAP6, PAP6, purple acid phosphatase 6 |
0.635 |
-0.672 |
| 49 |
261097_at |
AT1G62950
|
[leucine-rich repeat transmembrane protein kinase family protein] |
0.629 |
0.284 |
| 50 |
252907_at |
AT4G39650
|
GGT2, gamma-glutamyl transpeptidase 2 |
0.622 |
0.455 |
| 51 |
259405_at |
AT1G17744
|
unknown |
0.621 |
-0.335 |
| 52 |
262641_at |
AT1G62730
|
[Terpenoid synthases superfamily protein] |
0.614 |
-0.416 |
| 53 |
260947_at |
AT1G06020
|
[pfkB-like carbohydrate kinase family protein] |
0.609 |
-0.767 |
| 54 |
254152_at |
AT4G24410
|
unknown |
0.604 |
-0.518 |
| 55 |
254973_at |
AT4G10460
|
[copia-like retrotransposon family, has a 3.8e-291 P-value blast match to GB:CAA31653 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)] |
0.603 |
-0.201 |
| 56 |
248735_at |
AT5G48100
|
ATLAC15, AtTT10, LAC15, LACCASE-LIKE 15, TT10, TRANSPARENT TESTA 10 |
0.602 |
-1.165 |
| 57 |
251345_at |
AT3G60940
|
[Putative endonuclease or glycosyl hydrolase] |
0.598 |
0.124 |
| 58 |
256663_at |
AT3G12050
|
[Aha1 domain-containing protein] |
0.585 |
0.287 |
| 59 |
245058_at |
AT2G39790
|
[Mitochondrial glycoprotein family protein] |
0.584 |
-0.311 |
| 60 |
247647_at |
AT5G60010
|
[ferric reductase-like transmembrane component family protein] |
0.581 |
0.028 |
| 61 |
255916_at |
AT5G28550
|
unknown |
0.581 |
-0.702 |
| 62 |
253830_at |
AT4G27652
|
unknown |
0.580 |
0.554 |
| 63 |
256526_at |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
0.580 |
-1.125 |
| 64 |
265176_at |
AT1G23520
|
unknown |
0.579 |
-1.569 |
| 65 |
252176_at |
AT3G50720
|
[Protein kinase superfamily protein] |
0.575 |
-0.003 |
| 66 |
260662_at |
AT1G19540
|
[NmrA-like negative transcriptional regulator family protein] |
0.566 |
-0.541 |
| 67 |
247795_at |
AT5G58620
|
TZF9, tandem zinc finger protein 9 |
0.560 |
-0.388 |
| 68 |
257197_at |
AT3G23800
|
SBP3, selenium-binding protein 3 |
0.559 |
-0.565 |
| 69 |
264600_at |
AT1G04730
|
CTF18, CHROMOSOME TRANSMISSION FIDELITY 18 |
0.559 |
0.148 |
| 70 |
259084_at |
AT3G04800
|
ATTIM23-3, translocase inner membrane subunit 23-3, TIM23-3, translocase inner membrane subunit 23-3 |
0.556 |
0.410 |
| 71 |
262140_at |
AT1G52470
|
[alpha/beta-Hydrolases superfamily protein] |
0.554 |
-0.222 |
| 72 |
245243_at |
AT1G44414
|
unknown |
0.550 |
-0.575 |
| 73 |
257333_at |
ATMG01360
|
COX1, cytochrome oxidase |
0.550 |
-0.629 |
| 74 |
264150_at |
AT1G02090
|
ATCSN7, ARABIDOPSIS THALIANA COP9 SIGNALOSOME SUBUNIT 7, COP15, CONSTITUTIVE PHOTOMORPHOGENIC 15, CSN7, COP9 SIGNALOSOME SUBUNIT 7, FUS5, FUSCA 5 |
0.544 |
-0.018 |
| 75 |
253837_at |
AT4G27850
|
[Glycine-rich protein family] |
0.543 |
-0.525 |
| 76 |
261564_at |
AT1G01720
|
ANAC002, Arabidopsis NAC domain containing protein 2, ATAF1 |
0.541 |
0.078 |
| 77 |
259549_at |
AT1G35290
|
ALT1, acyl-lipid thioesterase 1 |
0.540 |
-0.273 |
| 78 |
258506_at |
AT3G06520
|
[agenet domain-containing protein] |
0.535 |
0.759 |
| 79 |
257322_at |
ATMG01180
|
ORF111B |
0.534 |
-1.119 |
| 80 |
254070_at |
AT4G25430
|
TRM23, TON1 Recruiting Motif 23 |
0.532 |
-1.121 |
| 81 |
258582_at |
AT3G04150
|
[RmlC-like cupins superfamily protein] |
0.530 |
-0.715 |
| 82 |
252455_at |
AT3G47140
|
[F-box associated ubiquitination effector family protein] |
0.522 |
-0.271 |
| 83 |
249389_at |
AT5G40100
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.518 |
-0.657 |
| 84 |
258830_at |
AT3G07090
|
[PPPDE putative thiol peptidase family protein] |
0.516 |
0.563 |
| 85 |
258872_at |
AT3G03260
|
HDG8, homeodomain GLABROUS 8 |
0.512 |
-0.424 |
| 86 |
266546_at |
AT2G35270
|
AHL21, AT-hook motif nuclear localized protein 21, GIK, GIANT KILLER |
0.511 |
-0.423 |
| 87 |
264710_at |
AT1G09790
|
COBL6, COBRA-like protein 6 precursor |
0.503 |
0.478 |
| 88 |
263546_at |
AT2G21550
|
[Bifunctional dihydrofolate reductase/thymidylate synthase] |
0.499 |
0.443 |
| 89 |
266754_at |
AT2G46980
|
ASY3, ASYNAPTIC 3, AtASY3, Arabidopsis thaliana ASYNAPTIC 3 |
0.498 |
-1.434 |
| 90 |
256416_at |
AT3G11050
|
ATFER2, ferritin 2, FER2, ferritin 2 |
0.497 |
-0.513 |
| 91 |
246733_at |
AT5G27660
|
DEG14, degradation of periplasmic proteins 14 |
0.495 |
0.993 |
| 92 |
265526_x_at |
AT2G06160
|
[gypsy-like retrotransposon family (Athila), has a 0. P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
0.489 |
-1.029 |
| 93 |
260349_at |
AT1G69400
|
[Transducin/WD40 repeat-like superfamily protein] |
0.488 |
-0.009 |
| 94 |
262543_at |
AT1G34245
|
EPF2, EPIDERMAL PATTERNING FACTOR 2 |
0.487 |
-0.276 |
| 95 |
254056_at |
AT4G25250
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.487 |
-0.050 |
| 96 |
256460_at |
AT1G36240
|
[Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein] |
0.484 |
-0.274 |
| 97 |
264331_at |
AT1G04130
|
AtTPR2, TPR2, tetratricopeptide repeat 2 |
0.483 |
-0.566 |
| 98 |
248045_at |
AT5G56030
|
AtHsp90.2, HEAT SHOCK PROTEIN 90.2, ERD8, EARLY-RESPONSIVE TO DEHYDRATION 8, HSP81-2, heat shock protein 81-2, HSP81.2, heat shock protein 81.2, HSP90.2, HEAT SHOCK PROTEIN 90.2 |
0.482 |
0.412 |
| 99 |
254931_at |
AT4G11460
|
CRK30, cysteine-rich RLK (RECEPTOR-like protein kinase) 30 |
0.480 |
-0.719 |
| 100 |
251947_at |
AT3G53550
|
[FBD-like domain family protein] |
0.478 |
-0.368 |
| 101 |
257691_at |
AT3G12660
|
FLA14, FASCICLIN-like arabinogalactan protein 14 precursor |
0.476 |
0.320 |
| 102 |
247002_at |
AT5G67320
|
HOS15, high expression of osmotically responsive genes 15 |
0.473 |
0.346 |
| 103 |
254753_at |
AT4G13190
|
[Protein kinase superfamily protein] |
0.471 |
-0.528 |
| 104 |
246254_at |
AT4G36450
|
ATMPK14, mitogen-activated protein kinase 14, MPK14, MPK14, mitogen-activated protein kinase 14 |
0.471 |
-0.929 |
| 105 |
259799_at |
AT1G72290
|
[Kunitz family trypsin and protease inhibitor protein] |
0.468 |
-0.344 |
| 106 |
263401_at |
AT2G04070
|
[MATE efflux family protein] |
0.466 |
-0.779 |
| 107 |
255316_at |
AT4G04170
|
[CACTA-like transposase family (Ptta/En/Spm), has a 2.3e-87 P-value blast match to At5g36655.1/81-333 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
0.465 |
-0.843 |
| 108 |
253683_at |
AT4G29650
|
[Cytidine/deoxycytidylate deaminase family protein] |
0.464 |
-0.892 |
| 109 |
261067_at |
AT1G07460
|
[Concanavalin A-like lectin family protein] |
0.463 |
-0.075 |
| 110 |
249858_at |
AT5G23000
|
ATMYB37, MYB37, myb domain protein 37, RAX1, REGULATOR OF AXILLARY MERISTEMS 1 |
0.463 |
-0.328 |
| 111 |
258191_at |
AT3G29100
|
ATVTI13, VTI13, vesicle transport V-snare 13 |
0.462 |
0.653 |
| 112 |
248275_at |
AT5G53520
|
ATOPT8, oligopeptide transporter 8, OPT8, oligopeptide transporter 8 |
0.460 |
-0.707 |
| 113 |
264554_at |
AT1G09510
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.459 |
0.186 |
| 114 |
261545_at |
AT1G63530
|
unknown |
0.458 |
-1.107 |
| 115 |
257029_at |
AT3G19240
|
[Vacuolar import/degradation, Vid27-related protein] |
0.457 |
0.368 |
| 116 |
255244_at |
AT4G05620
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.456 |
0.308 |
| 117 |
253483_at |
AT4G31910
|
BAT1, BR-related AcylTransferase1, PIZ, PIZZA |
0.455 |
-0.690 |
| 118 |
263489_at |
AT2G31830
|
5PTase14, inositol-polyphosphate 5-phosphatase 14 |
0.452 |
-0.508 |
| 119 |
246395_at |
AT1G58170
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.452 |
0.341 |
| 120 |
255730_at |
AT1G25460
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.451 |
-0.210 |
| 121 |
265500_at |
AT2G15750
|
unknown |
0.447 |
0.432 |
| 122 |
260608_at |
AT2G43870
|
[Pectin lyase-like superfamily protein] |
0.446 |
-0.438 |
| 123 |
254524_at |
AT4G20000
|
[VQ motif-containing protein] |
0.446 |
-0.245 |
| 124 |
265992_at |
AT2G24130
|
[Leucine-rich receptor-like protein kinase family protein] |
0.446 |
-0.587 |
| 125 |
252734_at |
AT3G43160
|
MEE38, maternal effect embryo arrest 38 |
0.445 |
-1.429 |
| 126 |
246203_at |
AT4G36610
|
[alpha/beta-Hydrolases superfamily protein] |
0.443 |
-0.498 |
| 127 |
259931_at |
AT1G34400
|
unknown |
0.441 |
-0.599 |
| 128 |
264814_at |
AT2G17900
|
ASHR1, ASH1-related 1, SDG37, SET domain group 37 |
0.440 |
0.221 |
| 129 |
251156_at |
AT3G63060
|
EDL3, EID1-like 3 |
0.440 |
-1.250 |
| 130 |
265852_at |
AT2G42350
|
[RING/U-box superfamily protein] |
0.439 |
-0.136 |
| 131 |
263469_at |
AT2G31910
|
ATCHX21, cation/H+ exchanger 21, CHX21, cation/H+ exchanger 21 |
0.439 |
-1.018 |
| 132 |
247531_at |
AT5G61550
|
[U-box domain-containing protein kinase family protein] |
0.437 |
-0.574 |
| 133 |
248245_at |
AT5G53190
|
AtSWEET3, SWEET3 |
0.436 |
-0.700 |
| 134 |
260658_at |
AT1G19410
|
[FBD / Leucine Rich Repeat domains containing protein] |
0.436 |
-0.619 |
| 135 |
254288_at |
AT4G22970
|
AESP, homolog of separase, ESP, EXTRA SPINDLE POLES, ESP, homolog of separase, RSW4, RADIALLY SWOLLEN 4 |
0.436 |
0.448 |
| 136 |
247290_at |
AT5G64450
|
unknown |
0.434 |
-0.327 |
| 137 |
250322_at |
AT5G12870
|
ATMYB46, MYB DOMAIN PROTEIN 46, MYB46, myb domain protein 46 |
0.433 |
-0.312 |
| 138 |
267222_at |
AT2G43880
|
[Pectin lyase-like superfamily protein] |
0.432 |
-0.379 |
| 139 |
248277_at |
AT5G52860
|
ABCG8, ATP-binding cassette G8 |
0.430 |
-0.370 |
| 140 |
258577_at |
AT3G04220
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.427 |
-0.356 |
| 141 |
264726_at |
AT1G22985
|
CRF7, cytokinin response factor 7 |
0.427 |
-0.322 |
| 142 |
247736_at |
AT5G59370
|
ACT4, actin 4 |
0.427 |
-0.639 |
| 143 |
267254_at |
AT2G23030
|
SNRK2-9, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-9, SNRK2.9, SNF1-related protein kinase 2.9 |
0.424 |
-0.396 |
| 144 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.423 |
-0.678 |
| 145 |
261752_at |
AT1G76290
|
[AMP-dependent synthetase and ligase family protein] |
0.421 |
-0.611 |
| 146 |
245228_at |
AT3G29810
|
COBL2, COBRA-like protein 2 precursor |
0.420 |
0.293 |
| 147 |
262580_at |
AT1G15330
|
[Cystathionine beta-synthase (CBS) protein] |
0.418 |
-0.502 |
| 148 |
248253_at |
AT5G53290
|
CRF3, cytokinin response factor 3 |
0.418 |
0.218 |
| 149 |
260824_at |
AT1G06720
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.418 |
-0.239 |
| 150 |
247871_at |
AT5G57530
|
AtXTH12, XTH12, xyloglucan endotransglucosylase/hydrolase 12 |
0.416 |
-0.165 |
| 151 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
0.416 |
-0.518 |
| 152 |
258754_at |
AT3G11920
|
[glutaredoxin-related] |
0.415 |
-0.578 |
| 153 |
247714_at |
AT5G59340
|
WOX2, WUSCHEL related homeobox 2 |
0.415 |
-1.193 |
| 154 |
265047_at |
AT1G51960
|
IQD27, IQ-domain 27 |
0.415 |
-0.539 |
| 155 |
262704_at |
AT1G16530
|
ASL9, ASYMMETRIC LEAVES 2-like 9, LBD3, LOB DOMAIN-CONTAINING PROTEIN 3 |
0.414 |
0.857 |
| 156 |
263543_at |
AT2G21610
|
ATPE11, A. THALIANA PECTINESTERASE 11, PE11, pectinesterase 11 |
0.414 |
0.549 |
| 157 |
251678_at |
AT3G56990
|
EDA7, embryo sac development arrest 7 |
0.414 |
-0.188 |
| 158 |
266773_at |
AT2G29040
|
[Exostosin family protein] |
0.414 |
-1.622 |
| 159 |
262865_at |
AT1G64930
|
CYP89A7, cytochrome P450, family 87, subfamily A, polypeptide 7 |
0.414 |
-1.230 |
| 160 |
249789_at |
AT5G24340
|
[3'-5' exonuclease domain-containing protein] |
0.413 |
1.056 |
| 161 |
249913_at |
AT5G22810
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.412 |
-0.313 |
| 162 |
263388_at |
AT2G11690
|
[CACTA-like transposase family (Tnp1/En/Spm), has a 4.8e-34 P-value blast match to ref|NP_189784.1| TNP1-related protein (Arabidopsis thaliana) (CACTA-element)] |
0.410 |
-0.658 |
| 163 |
251277_at |
AT3G61760
|
ADL1B, DYNAMIN-like 1B, DL1B, DYNAMIN-like 1B |
0.409 |
-0.597 |
| 164 |
250962_at |
AT5G02990
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.407 |
-0.164 |
| 165 |
259274_at |
AT3G01220
|
ATHB20, homeobox protein 20, HB20, homeobox protein 20 |
0.407 |
-0.468 |
| 166 |
258748_at |
AT3G05930
|
GLP8, germin-like protein 8 |
0.407 |
-0.134 |
| 167 |
258139_at |
AT3G24520
|
AT-HSFC1, HSFC1, heat shock transcription factor C1 |
0.405 |
0.093 |
| 168 |
262243_at |
AT1G48290
|
unknown |
0.402 |
-0.089 |
| 169 |
250396_at |
AT5G10970
|
[C2H2 and C2HC zinc fingers superfamily protein] |
0.400 |
-0.618 |
| 170 |
248770_at |
AT5G47740
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.399 |
-0.636 |
| 171 |
262004_at |
AT1G64480
|
CBL8, calcineurin B-like protein 8 |
0.399 |
-0.999 |
| 172 |
252019_at |
AT3G53040
|
[late embryogenesis abundant protein, putative / LEA protein, putative] |
0.398 |
-1.151 |
| 173 |
249770_at |
AT5G24110
|
ATWRKY30, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 30, WRKY30, WRKY DNA-binding protein 30 |
0.398 |
-0.314 |
| 174 |
249326_at |
AT5G40840
|
AtRAD21.1, Sister chromatid cohesion 1 (SCC1) protein homolog 2, SYN2 |
0.395 |
0.297 |
| 175 |
245904_at |
AT5G11110
|
ATSPS2F, sucrose phosphate synthase 2F, KNS2, KAONASHI 2, SPS1, SUCROSE PHOSPHATE SYNTHASE 1, SPS2F, sucrose phosphate synthase 2F, SPSA2, sucrose-phosphate synthase A2 |
0.395 |
-0.755 |
| 176 |
245921_at |
AT5G28800
|
unknown |
0.394 |
-0.862 |
| 177 |
250841_at |
AT5G04610
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.393 |
-0.749 |
| 178 |
259251_at |
AT3G07600
|
[Heavy metal transport/detoxification superfamily protein ] |
0.392 |
-0.983 |
| 179 |
261305_at |
AT1G48470
|
GLN1;5, glutamine synthetase 1;5 |
0.390 |
-0.806 |
| 180 |
260424_at |
AT1G72460
|
[Leucine-rich repeat protein kinase family protein] |
0.390 |
-0.239 |
| 181 |
245393_at |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
0.389 |
0.266 |
| 182 |
260600_at |
AT1G55950
|
[DNA-binding storekeeper protein-related transcriptional regulator] |
0.389 |
0.162 |
| 183 |
260344_at |
AT1G69240
|
ATMES15, ARABIDOPSIS THALIANA METHYL ESTERASE 15, MES15, methyl esterase 15, RHS9, ROOT HAIR SPECIFIC 9 |
0.388 |
0.240 |
| 184 |
247704_at |
AT5G59510
|
DVL18, DEVIL 18, RTFL5, ROTUNDIFOLIA like 5 |
0.387 |
-0.533 |
| 185 |
245335_at |
AT4G16160
|
ATOEP16-2, ATOEP16-S |
0.387 |
-0.886 |
| 186 |
248774_at |
AT5G47830
|
unknown |
0.386 |
0.265 |
| 187 |
261700_at |
AT1G32690
|
unknown |
0.386 |
0.685 |
| 188 |
259580_at |
AT1G28030
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.384 |
0.000 |
| 189 |
267472_at |
AT2G02850
|
ARPN, plantacyanin |
0.383 |
0.320 |
| 190 |
245241_at |
AT1G44478
|
[Cyclophilin] |
0.383 |
-1.065 |
| 191 |
249458_at |
AT5G39560
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.382 |
0.285 |
| 192 |
258503_at |
AT3G02500
|
unknown |
0.381 |
-0.857 |
| 193 |
253771_at |
AT4G28430
|
[Reticulon family protein] |
0.381 |
-0.843 |
| 194 |
253036_at |
AT4G38340
|
NLP3, NIN-like protein 3 |
0.381 |
-0.609 |
| 195 |
249907_at |
AT5G22750
|
RAD5, RAD5A |
0.380 |
-0.321 |
| 196 |
263861_at |
AT2G04560
|
AtLpxB, LpxB, lipid X B |
0.380 |
-0.723 |
| 197 |
246686_at |
AT5G33380
|
unknown |
0.379 |
-0.003 |
| 198 |
256081_at |
AT1G20700
|
ATWOX14, WUSCHEL RELATED HOMEOBOX 14, WOX14, WUSCHEL related homeobox 14 |
0.378 |
-0.809 |
| 199 |
245663_at |
AT1G28170
|
SOT7, sulphotransferase 7 |
0.378 |
-0.829 |
| 200 |
252653_at |
AT3G44730
|
AtKIN14h, ATKP1, ARABIDOPSIS KINESIN-LIKE PROTEIN 1, KP1, kinesin-like protein 1 |
0.378 |
-0.599 |
| 201 |
264862_at |
AT1G24330
|
[ARM repeat superfamily protein] |
0.377 |
-1.084 |
| 202 |
245460_at |
AT4G16990
|
RLM3, RESISTANCE TO LEPTOSPHAERIA MACULANS 3 |
-1.169 |
0.995 |
| 203 |
267367_at |
AT2G44210
|
unknown |
-1.167 |
0.720 |
| 204 |
258897_at |
AT3G05730
|
[LOCATED IN: endomembrane system] |
-1.148 |
0.922 |
| 205 |
251746_at |
AT3G56060
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
-1.056 |
0.162 |
| 206 |
252206_at |
AT3G50360
|
ATCEN2, centrin2, CEN1, CENTRIN 1, CEN2, centrin2 |
-1.039 |
0.939 |
| 207 |
261822_at |
AT1G11380
|
[PLAC8 family protein] |
-1.023 |
-0.224 |
| 208 |
264316_at |
AT1G70330
|
ENT1, equilibrative nucleotide transporter 1, ENT1,AT, equilibrative nucleotide transporter 1 |
-0.994 |
0.525 |
| 209 |
245242_at |
AT1G44446
|
ATCAO, ARABIDOPSIS THALIANA CHLOROPHYLL A OXYGENASE, CAO, CHLOROPHYLL A OXYGENASE, CH1, CHLORINA 1 |
-0.986 |
0.974 |
| 210 |
257269_at |
AT3G17440
|
ATNPSN13, NPSN13, novel plant snare 13 |
-0.971 |
0.340 |
| 211 |
255913_at |
AT1G66980
|
GDPDL2, Glycerophosphodiester phosphodiesterase (GDPD) like 2, SNC4, suppressor of npr1-1 constitutive 4 |
-0.968 |
-0.626 |
| 212 |
261453_at |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
-0.967 |
0.098 |
| 213 |
266658_at |
AT2G25735
|
unknown |
-0.947 |
0.018 |
| 214 |
257831_at |
AT3G26710
|
CCB1, cofactor assembly of complex C |
-0.947 |
1.147 |
| 215 |
261141_at |
AT1G19740
|
[ATP-dependent protease La (LON) domain protein] |
-0.941 |
0.659 |
| 216 |
249847_at |
AT5G23210
|
SCPL34, serine carboxypeptidase-like 34 |
-0.941 |
0.095 |
| 217 |
259658_at |
AT1G55370
|
NDF5, NDH-dependent cyclic electron flow 5 |
-0.936 |
0.397 |
| 218 |
254040_at |
AT4G25900
|
[Galactose mutarotase-like superfamily protein] |
-0.931 |
0.022 |
| 219 |
249291_at |
AT5G41210
|
ATGSTT1, glutathione S-transferase THETA 1, GST10, GSTT1, glutathione S-transferase THETA 1 |
-0.929 |
0.656 |
| 220 |
255657_at |
AT4G00810
|
[60S acidic ribosomal protein family] |
-0.928 |
1.419 |
| 221 |
249583_at |
AT5G37770
|
CML24, CALMODULIN-LIKE 24, TCH2, TOUCH 2 |
-0.923 |
0.570 |
| 222 |
264238_at |
AT1G54740
|
unknown |
-0.919 |
-0.612 |
| 223 |
262569_at |
AT1G15180
|
[MATE efflux family protein] |
-0.912 |
0.481 |
| 224 |
264955_at |
AT1G76920
|
[F-box family protein] |
-0.907 |
-0.079 |
| 225 |
255046_at |
AT4G09650
|
ATPD, ATP synthase delta-subunit gene, PDE332, PIGMENT DEFECTIVE 332 |
-0.899 |
0.799 |
| 226 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
-0.890 |
0.185 |
| 227 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-0.886 |
-0.703 |
| 228 |
252533_at |
AT3G46110
|
[LOCATED IN: plasma membrane] |
-0.882 |
-0.481 |
| 229 |
267645_at |
AT2G32860
|
BGLU33, beta glucosidase 33 |
-0.881 |
0.162 |
| 230 |
251507_at |
AT3G59080
|
[Eukaryotic aspartyl protease family protein] |
-0.880 |
-0.262 |
| 231 |
252573_at |
AT3G45260
|
[C2H2-like zinc finger protein] |
-0.876 |
0.278 |
| 232 |
247440_at |
AT5G62680
|
AtNPF2.11, GTR2, GLUCOSINOLATE TRANSPORTER-2, NPF2.11, NRT1/ PTR family 2.11 |
-0.874 |
-0.150 |
| 233 |
255645_at |
AT4G00880
|
SAUR31, SMALL AUXIN UPREGULATED RNA 31 |
-0.872 |
0.665 |
| 234 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
-0.869 |
1.155 |
| 235 |
254924_at |
AT4G11330
|
ATMPK5, MAP kinase 5, MPK5, MAP kinase 5 |
-0.864 |
-0.205 |
| 236 |
248950_at |
AT5G45390
|
CLPP4, CLP protease P4, NCLPP4, NUCLEAR-ENCODED CLP PROTEASE P4 |
-0.863 |
1.772 |
| 237 |
262945_at |
AT1G79510
|
[Uncharacterized conserved protein (DUF2358)] |
-0.861 |
0.333 |
| 238 |
262768_at |
AT1G12990
|
[beta-1,4-N-acetylglucosaminyltransferase family protein] |
-0.860 |
0.253 |
| 239 |
260870_at |
AT1G43890
|
ATRAB-C1, ARABIDOPSIS RAB GTPASE HOMOLOG C1, ATRAB18, ARABIDOPSIS RAB GTPASE HOMOLOG B18, ATRABC1, ARABIDOPSIS RAB GTPASE HOMOLOG C1, RAB18, RAB GTPASE HOMOLOG B18, RAB18-1, RAB GTPASE HOMOLOG 18-1, RABC1, RAB GTPASE HOMOLOG C1 |
-0.858 |
0.485 |
| 240 |
267578_at |
AT2G30695
|
unknown |
-0.852 |
0.223 |
| 241 |
262951_at |
AT1G75500
|
UMAMIT5, Usually multiple acids move in and out Transporters 5, WAT1, Walls Are Thin 1 |
-0.847 |
0.395 |
| 242 |
257717_at |
AT3G18390
|
EMB1865, embryo defective 1865 |
-0.844 |
0.837 |
| 243 |
265918_at |
AT2G15090
|
KCS8, 3-ketoacyl-CoA synthase 8 |
-0.842 |
0.211 |
| 244 |
256456_at |
AT1G75180
|
[Erythronate-4-phosphate dehydrogenase family protein] |
-0.840 |
0.762 |
| 245 |
245877_at |
AT1G26220
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.839 |
-0.552 |
| 246 |
248094_at |
AT5G55220
|
[trigger factor type chaperone family protein] |
-0.837 |
0.432 |
| 247 |
246730_at |
AT5G28060
|
[Ribosomal protein S24e family protein] |
-0.834 |
1.616 |
| 248 |
256021_at |
AT1G58270
|
ZW9 |
-0.831 |
-0.789 |
| 249 |
257913_at |
AT3G25540
|
LOH1, LAG One Homologue 1 |
-0.826 |
0.187 |
| 250 |
246143_at |
AT5G19980
|
GONST4, golgi nucleotide sugar transporter 4 |
-0.823 |
0.278 |
| 251 |
260567_at |
AT2G43820
|
ATSAGT1, Arabidopsis thaliana salicylic acid glucosyltransferase 1, GT, SAGT1, salicylic acid glucosyltransferase 1, SGT1, UDP-glucose:salicylic acid glucosyltransferase 1, UGT74F2, UDP-glucosyltransferase 74F2 |
-0.821 |
0.119 |
| 252 |
250549_at |
AT5G07860
|
[HXXXD-type acyl-transferase family protein] |
-0.819 |
0.958 |
| 253 |
250355_at |
AT5G11700
|
[LOCATED IN: vacuole] |
-0.814 |
0.281 |
| 254 |
254815_at |
AT4G12420
|
SKU5 |
-0.811 |
0.617 |
| 255 |
256570_at |
AT3G19540
|
unknown |
-0.811 |
-0.071 |
| 256 |
245324_at |
AT4G17260
|
[Lactate/malate dehydrogenase family protein] |
-0.811 |
-0.147 |
| 257 |
266813_at |
AT2G44920
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.808 |
0.748 |
| 258 |
250881_at |
AT5G04080
|
unknown |
-0.806 |
-0.908 |
| 259 |
254077_at |
AT4G25640
|
ATDTX35, detoxifying efflux carrier 35, DTX35, detoxifying efflux carrier 35, FFT, FLOWER FLAVONOID TRANSPORTER |
-0.799 |
0.536 |
| 260 |
254860_at |
AT4G12110
|
ATSMO1-1, SMO1-1, sterol-4alpha-methyl oxidase 1-1 |
-0.797 |
0.190 |
| 261 |
249109_at |
AT5G43700
|
ATAUX2-11, AUXIN INDUCIBLE 2-11, IAA4, indole-3-acetic acid inducible 4 |
-0.793 |
0.748 |
| 262 |
263137_at |
AT1G78660
|
ATGGH1, gamma-glutamyl hydrolase 1, GGH1, gamma-glutamyl hydrolase 1 |
-0.793 |
0.022 |
| 263 |
246929_at |
AT5G25210
|
unknown |
-0.790 |
-1.086 |
| 264 |
257894_at |
AT3G17100
|
AIF3, ATBS1 Interacting Factor 3 |
-0.790 |
0.537 |
| 265 |
245768_at |
AT1G33590
|
[Leucine-rich repeat (LRR) family protein] |
-0.788 |
0.748 |
| 266 |
257793_at |
AT3G26960
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.788 |
-0.077 |
| 267 |
254638_at |
AT4G18740
|
[Rho termination factor] |
-0.785 |
0.214 |
| 268 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
-0.784 |
-0.317 |
| 269 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
-0.780 |
-0.596 |
| 270 |
253571_at |
AT4G31000
|
[Calmodulin-binding protein] |
-0.780 |
-0.349 |
| 271 |
251557_at |
AT3G58730
|
[vacuolar ATP synthase subunit D (VATD) / V-ATPase D subunit / vacuolar proton pump D subunit (VATPD)] |
-0.779 |
0.523 |
| 272 |
250939_at |
AT5G03040
|
iqd2, IQ-domain 2 |
-0.776 |
0.003 |
| 273 |
257260_at |
AT3G22104
|
[Phototropic-responsive NPH3 family protein] |
-0.775 |
0.344 |
| 274 |
267220_at |
AT2G02500
|
ATMEPCT, ISPD, MCT, 2-C-METHYL-D-ERYTHRITOL 4-PHOSPHATE CYTIDYLTRANSFERASE |
-0.774 |
0.344 |
| 275 |
267262_at |
AT2G22990
|
SCPL8, SERINE CARBOXYPEPTIDASE-LIKE 8, SNG1, sinapoylglucose 1 |
-0.774 |
0.164 |
| 276 |
246194_at |
AT4G37000
|
ACD2, ACCELERATED CELL DEATH 2, ATRCCR, ARABIDOPSIS THALIANA RED CHLOROPHYLL CATABOLITE REDUCTASE |
-0.768 |
0.257 |
| 277 |
267268_at |
AT2G02570
|
[nucleic acid binding] |
-0.765 |
0.105 |
| 278 |
262005_at |
AT1G64550
|
ABCF3, ATP-binding cassette F3, AtABCF3, AtGCN20, ATGCN3, general control non-repressible 3, GCN20, general control non-repressible 20, GCN3, general control non-repressible 3, SCORD5, susceptible to coronatine-deficient Pst DC3000 5 |
-0.761 |
0.814 |
| 279 |
253465_at |
AT4G32120
|
[Galactosyltransferase family protein] |
-0.759 |
0.367 |
| 280 |
245716_at |
AT5G08740
|
NDC1, NAD(P)H dehydrogenase C1 |
-0.759 |
0.834 |
| 281 |
262871_at |
AT1G65010
|
[INVOLVED IN: flower development] |
-0.758 |
-0.574 |
| 282 |
260489_at |
AT1G51610
|
[Cation efflux family protein] |
-0.756 |
0.989 |
| 283 |
264980_at |
AT1G27190
|
[Leucine-rich repeat protein kinase family protein] |
-0.755 |
0.156 |
| 284 |
266803_at |
AT2G28930
|
APK1B, protein kinase 1B, PK1B, protein kinase 1B |
-0.755 |
-0.423 |
| 285 |
246034_at |
AT5G08350
|
[GRAM domain-containing protein / ABA-responsive protein-related] |
-0.755 |
0.599 |
| 286 |
257621_at |
AT3G20410
|
CPK9, calmodulin-domain protein kinase 9 |
-0.754 |
0.299 |
| 287 |
261981_at |
AT1G33811
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.753 |
0.391 |
| 288 |
253440_at |
AT4G32570
|
TIFY8, TIFY domain protein 8 |
-0.751 |
0.974 |
| 289 |
257202_at |
AT3G23750
|
BARK1, BAK1-Associating Receptor-Like Kinase 1 |
-0.746 |
0.776 |
| 290 |
263480_at |
AT2G04032
|
ZIP7, zinc transporter 7 precursor |
-0.746 |
0.593 |
| 291 |
259523_at |
AT1G12500
|
[Nucleotide-sugar transporter family protein] |
-0.745 |
1.140 |
| 292 |
245816_at |
AT1G26210
|
ATSOFL1, SOB FIVE-LIKE 1, SOFL1, SOB five-like 1 |
-0.744 |
0.039 |
| 293 |
250996_at |
AT5G02530
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.743 |
1.206 |
| 294 |
254998_at |
AT4G09760
|
[Protein kinase superfamily protein] |
-0.743 |
-0.094 |
| 295 |
260020_at |
AT1G29990
|
PFD6, prefoldin 6 |
-0.743 |
0.842 |
| 296 |
266964_at |
AT2G39480
|
ABCB6, ATP-binding cassette B6, PGP6, P-glycoprotein 6 |
-0.742 |
-0.499 |
| 297 |
260055_at |
AT1G78150
|
unknown |
-0.742 |
1.276 |
| 298 |
247552_at |
AT5G60920
|
COB, COBRA |
-0.740 |
0.106 |
| 299 |
251314_at |
AT3G61180
|
[RING/U-box superfamily protein] |
-0.737 |
0.096 |
| 300 |
258113_at |
AT3G14650
|
CYP72A11, cytochrome P450, family 72, subfamily A, polypeptide 11 |
-0.735 |
-0.224 |
| 301 |
260636_at |
AT1G62430
|
ATCDS1, CDP-diacylglycerol synthase 1, CDS1, CDP-diacylglycerol synthase 1 |
-0.734 |
0.001 |
| 302 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
-0.731 |
-0.785 |
| 303 |
260126_at |
AT1G36370
|
SHM7, serine hydroxymethyltransferase 7 |
-0.728 |
0.025 |
| 304 |
250847_at |
AT5G04480
|
[UDP-Glycosyltransferase superfamily protein] |
-0.727 |
0.793 |
| 305 |
253393_at |
AT4G32690
|
ATGLB3, ARABIDOPSIS HEMOGLOBIN 3, GLB3, hemoglobin 3 |
-0.725 |
-0.691 |
| 306 |
258048_at |
AT3G16290
|
EMB2083, embryo defective 2083 |
-0.723 |
0.347 |
| 307 |
255899_at |
AT1G17970
|
[RING/U-box superfamily protein] |
-0.720 |
0.086 |
| 308 |
259681_at |
AT1G77760
|
GNR1, NIA1, nitrate reductase 1, NR1, NITRATE REDUCTASE 1 |
-0.716 |
1.038 |
| 309 |
267617_at |
AT2G26670
|
ATHO1, ARABIDOPSIS THALIANA HEME OXYGENASE 1, GUN2, GENOMES UNCOUPLED 2, HO1, HEME OXYGENASE 1, HY1, HEME OXYGENASE 1, HY6, HEME OXYGENASE 6, TED4, REVERSAL OF THE DET PHENOTYPE 4 |
-0.715 |
0.402 |
| 310 |
247994_at |
AT5G56140
|
[RNA-binding KH domain-containing protein] |
-0.715 |
0.590 |
| 311 |
264246_at |
AT1G60140
|
ATTPS10, trehalose phosphate synthase, TPS10, trehalose phosphate synthase, TPS10, TREHALOSE PHOSPHATE SYNTHASE 10 |
-0.713 |
0.423 |
| 312 |
258608_at |
AT3G03020
|
unknown |
-0.709 |
0.461 |
| 313 |
266705_at |
AT2G19750
|
[Ribosomal protein S30 family protein] |
-0.708 |
0.444 |
| 314 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-0.708 |
1.354 |
| 315 |
254093_at |
AT4G25110
|
AtMC2, metacaspase 2, AtMCP1c, metacaspase 1c, MC2, metacaspase 2, MCP1c, metacaspase 1c |
-0.706 |
-0.578 |
| 316 |
266231_at |
AT2G02220
|
ATPSKR1, PHYTOSULFOKIN RECEPTOR 1, PSKR1, phytosulfokin receptor 1 |
-0.703 |
-0.282 |
| 317 |
250470_at |
AT5G10160
|
[Thioesterase superfamily protein] |
-0.703 |
0.323 |
| 318 |
247924_at |
AT5G57655
|
[xylose isomerase family protein] |
-0.703 |
-0.159 |
| 319 |
248942_at |
AT5G45480
|
unknown |
-0.699 |
-0.059 |
| 320 |
264698_at |
AT1G70200
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.696 |
0.106 |
| 321 |
247316_at |
AT5G64030
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.695 |
0.202 |
| 322 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.695 |
1.013 |
| 323 |
258060_at |
AT3G26030
|
ATB' DELTA, serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B prime delta |
-0.694 |
0.473 |
| 324 |
252066_at |
AT3G51550
|
FER, FERONIA |
-0.694 |
1.360 |
| 325 |
249930_at |
AT5G22360
|
ATVAMP714, vesicle-associated membrane protein 714, VAMP714, VAMP714, vesicle-associated membrane protein 714 |
-0.693 |
0.035 |
| 326 |
245027_at |
AT2G26550
|
HO2, heme oxygenase 2 |
-0.692 |
-0.194 |
| 327 |
257459_at |
AT1G24040
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.692 |
0.793 |
| 328 |
249473_at |
AT5G39340
|
AHP3, histidine-containing phosphotransmitter 3, ATHP2, ARABIDOPSIS THALIANA HISTIDINE-CONTAINING PHOSPHOTRANSMITTER 2 |
-0.691 |
0.373 |
| 329 |
251738_at |
AT3G56150
|
ATEIF3C-1, ATTIF3C1, EIF3C, eukaryotic translation initiation factor 3C, EIF3C-1, TIF3C1 |
-0.691 |
0.875 |
| 330 |
248303_at |
AT5G53170
|
FTSH11, FTSH protease 11 |
-0.690 |
0.790 |
| 331 |
263165_at |
AT1G03060
|
SPI, SPIRRIG |
-0.689 |
0.768 |
| 332 |
259010_at |
AT3G07340
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.689 |
-0.795 |
| 333 |
263606_at |
AT2G16280
|
KCS9, 3-ketoacyl-CoA synthase 9 |
-0.689 |
0.394 |
| 334 |
257971_at |
AT3G27530
|
GC6, golgin candidate 6, MAG4, MAIGO 4 |
-0.688 |
0.090 |
| 335 |
252075_at |
AT3G51730
|
[saposin B domain-containing protein] |
-0.684 |
0.488 |
| 336 |
254505_at |
AT4G19985
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.683 |
0.296 |
| 337 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
-0.683 |
0.231 |
| 338 |
255852_at |
AT1G66970
|
GDPDL1, Glycerophosphodiester phosphodiesterase (GDPD) like 1, SVL2, SHV3-like 2 |
-0.683 |
1.441 |
| 339 |
256015_at |
AT1G19150
|
LHCA2*1, Lhca6, photosystem I light harvesting complex gene 6 |
-0.682 |
0.942 |
| 340 |
251260_at |
AT3G62130
|
AtLCD, LCD, L-cysteine desulfhydrase |
-0.681 |
0.700 |
| 341 |
264910_at |
AT1G61100
|
[disease resistance protein (TIR class), putative] |
-0.680 |
1.107 |
| 342 |
252290_at |
AT3G49140
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.680 |
0.749 |
| 343 |
263555_at |
AT2G16460
|
unknown |
-0.677 |
0.054 |
| 344 |
261375_at |
AT1G53160
|
FTM6, FLORAL TRANSITION AT THE MERISTEM6, SPL4, squamosa promoter binding protein-like 4 |
-0.675 |
-0.501 |
| 345 |
247929_at |
AT5G57330
|
[Galactose mutarotase-like superfamily protein] |
-0.674 |
-0.141 |
| 346 |
258472_at |
AT3G06080
|
TBL10, TRICHOME BIREFRINGENCE-LIKE 10 |
-0.673 |
0.410 |
| 347 |
260571_at |
AT2G43790
|
ATMAPK6, MAP kinase 6, ATMPK6, MAP kinase 6, MAPK6, MAP kinase 6, MPK6, MAP kinase 6 |
-0.672 |
1.116 |
| 348 |
254416_at |
AT4G21380
|
ARK3, receptor kinase 3, RK3, receptor kinase 3 |
-0.671 |
0.051 |
| 349 |
258677_at |
AT3G08730
|
ATPK1, ARABIDOPSIS THALIANA PROTEIN-SERINE KINASE 1, ATPK6, ARABIDOPSIS THALIANA PROTEIN-SERINE KINASE 6, ATS6K1, PK1, protein-serine kinase 1, PK6, ROTEIN-SERINE KINASE 6, S6K1, P70 RIBOSOMAL S6 KINASE |
-0.671 |
1.685 |
| 350 |
256524_at |
AT1G66200
|
ATGSR2, glutamine synthase clone F11, GLN1;2, glutamine synthetase 1;2, GSR2, glutamine synthase clone F11 |
-0.670 |
0.682 |
| 351 |
260410_at |
AT1G69870
|
AtNPF2.13, NPF2.13, NRT1/ PTR family 2.13, NRT1.7, nitrate transporter 1.7 |
-0.668 |
0.900 |
| 352 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
-0.668 |
-0.078 |
| 353 |
265724_at |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
-0.667 |
1.197 |
| 354 |
260142_at |
AT1G71900
|
unknown |
-0.666 |
-0.019 |
| 355 |
265700_at |
AT2G03470
|
[ELM2 domain-containing protein] |
-0.664 |
0.729 |
| 356 |
266322_at |
AT2G46690
|
SAUR32, SMALL AUXIN UPREGULATED RNA 32 |
-0.664 |
0.602 |
| 357 |
266604_at |
AT2G46030
|
UBC6, ubiquitin-conjugating enzyme 6 |
-0.664 |
0.755 |
| 358 |
259017_at |
AT3G07310
|
unknown |
-0.662 |
0.628 |
| 359 |
264566_at |
AT1G05270
|
[TraB family protein] |
-0.662 |
0.733 |
| 360 |
258706_at |
AT3G09570
|
[Lung seven transmembrane receptor family protein] |
-0.660 |
0.977 |
| 361 |
245321_at |
AT4G15545
|
unknown |
-0.659 |
0.205 |
| 362 |
259769_at |
AT1G29400
|
AML5, MEI2-like protein 5, ML5, MEI2-like protein 5 |
-0.659 |
1.254 |
| 363 |
266635_at |
AT2G35470
|
unknown |
-0.658 |
0.085 |
| 364 |
258958_at |
AT3G01390
|
AVMA10, VMA10, vacuolar membrane ATPase 10 |
-0.658 |
1.321 |
| 365 |
246492_at |
AT5G16140
|
[Peptidyl-tRNA hydrolase family protein] |
-0.657 |
1.136 |
| 366 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
-0.655 |
0.104 |
| 367 |
257144_at |
AT3G27300
|
G6PD5, glucose-6-phosphate dehydrogenase 5 |
-0.655 |
0.542 |
| 368 |
256527_at |
AT1G66100
|
[Plant thionin] |
-0.655 |
-1.308 |
| 369 |
246995_at |
AT5G67470
|
ATFH6, ARABIDOPSIS FORMIN HOMOLOG 6, FH6, formin homolog 6 |
-0.654 |
-0.159 |
| 370 |
246555_at |
AT5G15470
|
GAUT14, galacturonosyltransferase 14 |
-0.652 |
0.104 |
| 371 |
253162_at |
AT4G35630
|
PSAT1, phosphoserine aminotransferase 1 |
-0.652 |
0.650 |
| 372 |
250527_at |
AT5G08590
|
ASK2, ARABIDOPSIS SERINE/THREONINE KINASE 2, ASK2, ARABIDOPSIS SKP1-LIKE1, SNRK2-1, SNRK2.1, SNF1-related protein kinase 2.1, SRK2G |
-0.651 |
1.344 |
| 373 |
265595_at |
AT2G20140
|
RPT2b, regulatory particle AAA-ATPase 2b |
-0.650 |
0.560 |
| 374 |
255232_at |
AT4G05330
|
AGD13, ARF-GAP domain 13 |
-0.648 |
-1.070 |
| 375 |
248541_at |
AT5G50180
|
[Protein kinase superfamily protein] |
-0.647 |
0.603 |
| 376 |
257909_at |
AT3G25480
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.646 |
0.575 |
| 377 |
247607_at |
AT5G60960
|
PNM1, PPR protein localized to the nucleus and mitochondria 1 |
-0.646 |
-0.199 |
| 378 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
-0.644 |
0.340 |
| 379 |
250210_at |
AT5G13890
|
unknown |
-0.642 |
0.646 |
| 380 |
263904_at |
AT2G36380
|
ABCG34, ATP-binding cassette G34, ATPDR6, PLEIOTROPIC DRUG RESISTANCE 6, PDR6, pleiotropic drug resistance 6 |
-0.641 |
0.330 |
| 381 |
264705_at |
AT1G09620
|
[ATP binding] |
-0.641 |
0.246 |
| 382 |
259677_at |
AT1G77740
|
PIP5K2, phosphatidylinositol-4-phosphate 5-kinase 2 |
-0.641 |
0.390 |
| 383 |
245417_at |
AT4G17360
|
[Formyl transferase] |
-0.637 |
0.982 |
| 384 |
261570_at |
AT1G01120
|
KCS1, 3-ketoacyl-CoA synthase 1 |
-0.636 |
0.358 |
| 385 |
250132_at |
AT5G16560
|
KAN, KANADI, KAN1, KANADI 1 |
-0.635 |
-0.539 |
| 386 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
-0.634 |
0.225 |
| 387 |
251396_at |
AT3G60750
|
AtTKL1, TKL1, transketolase 1 |
-0.633 |
0.958 |
| 388 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
-0.633 |
-0.310 |
| 389 |
252343_at |
AT3G48610
|
NPC6, non-specific phospholipase C6 |
-0.633 |
-0.121 |
| 390 |
247114_at |
AT5G65910
|
[BSD domain-containing protein] |
-0.632 |
-0.191 |
| 391 |
255786_at |
AT1G19670
|
ATCLH1, chlorophyllase 1, ATHCOR1, CORONATINE-INDUCED PROTEIN 1, CLH1, chlorophyllase 1, CORI1, CORONATINE-INDUCED PROTEIN 1 |
-0.632 |
0.515 |
| 392 |
266279_at |
AT2G29290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.632 |
1.681 |
| 393 |
246417_at |
AT5G16990
|
[Zinc-binding dehydrogenase family protein] |
-0.631 |
0.841 |
| 394 |
247601_at |
AT5G60850
|
OBP4, OBF binding protein 4 |
-0.629 |
0.584 |
| 395 |
246031_at |
AT5G21160
|
AtLARP1a, La related protein 1a, LARP1a, La related protein 1a |
-0.628 |
0.094 |
| 396 |
253284_at |
AT4G34150
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.628 |
0.583 |
| 397 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.626 |
0.550 |
| 398 |
257759_at |
AT3G23070
|
ATCFM3A, CFM3A, CRM family member 3A |
-0.626 |
0.027 |
| 399 |
246033_at |
AT5G08280
|
HEMC, hydroxymethylbilane synthase, RUG1, RUGOSA 1 |
-0.625 |
1.201 |
| 400 |
259765_at |
AT1G64370
|
unknown |
-0.625 |
-0.250 |
| 401 |
265962_at |
AT2G37460
|
UMAMIT12, Usually multiple acids move in and out Transporters 12 |
-0.624 |
0.087 |