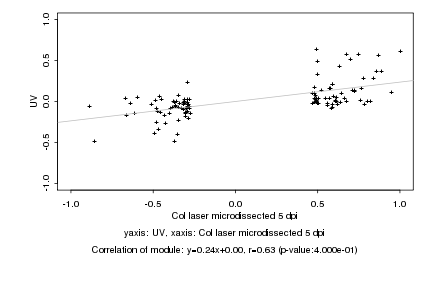
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Col laser microdissected 5 dpi |
Log2 signal ratio
UV |
| 1 |
260116_at |
AT1G33960
|
AIG1, AVRRPT2-INDUCED GENE 1 |
1.001 |
0.617 |
| 2 |
245076_at |
AT2G23170
|
GH3.3 |
0.943 |
0.115 |
| 3 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.885 |
0.370 |
| 4 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
0.869 |
0.566 |
| 5 |
253796_at |
AT4G28460
|
unknown |
0.852 |
0.378 |
| 6 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
0.834 |
0.292 |
| 7 |
248588_at |
AT5G49540
|
[Rab5-interacting family protein] |
0.815 |
0.012 |
| 8 |
263181_at |
AT1G05720
|
[selenoprotein family protein] |
0.802 |
0.012 |
| 9 |
250398_at |
AT5G11000
|
unknown |
0.782 |
-0.025 |
| 10 |
251422_at |
AT3G60540
|
[Preprotein translocase Sec, Sec61-beta subunit protein] |
0.773 |
0.287 |
| 11 |
246098_at |
AT5G20400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.766 |
0.160 |
| 12 |
262426_s_at |
AT1G47640
|
unknown |
0.760 |
0.024 |
| 13 |
248321_at |
AT5G52740
|
[Copper transport protein family] |
0.748 |
0.577 |
| 14 |
246327_at |
AT1G16670
|
[Protein kinase superfamily protein] |
0.723 |
0.136 |
| 15 |
254166_at |
AT4G24190
|
AtHsp90-7, HEAT SHOCK PROTEIN 90-7, AtHsp90.7, HEAT SHOCK PROTEIN 90.7, HSP90.7, HEAT SHOCK PROTEIN 90.7, SHD, SHEPHERD |
0.719 |
0.132 |
| 16 |
250738_at |
AT5G05730
|
AMT1, A-METHYL TRYPTOPHAN RESISTANT 1, ASA1, anthranilate synthase alpha subunit 1, JDL1, JASMONATE-INDUCED DEFECTIVE LATERAL ROOT 1, TRP5, TRYPTOPHAN BIOSYNTHESIS 5, WEI2, WEAK ETHYLENE INSENSITIVE 2 |
0.710 |
0.137 |
| 17 |
256596_at |
AT3G28540
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.698 |
0.518 |
| 18 |
258140_at |
AT3G24503
|
ALDH1A, aldehyde dehydrogenase 1A, ALDH2C4, aldehyde dehydrogenase 2C4, REF1, REDUCED EPIDERMAL FLUORESCENCE1 |
0.675 |
0.007 |
| 19 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
0.673 |
0.580 |
| 20 |
254429_at |
AT4G21105
|
[cytochrome-c oxidases] |
0.662 |
0.047 |
| 21 |
263466_at |
AT2G31740
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.644 |
0.107 |
| 22 |
263046_at |
AT2G05380
|
GRP3S, glycine-rich protein 3 short isoform |
0.634 |
-0.005 |
| 23 |
264434_at |
AT1G10340
|
[Ankyrin repeat family protein] |
0.628 |
0.431 |
| 24 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
0.617 |
-0.026 |
| 25 |
260724_at |
AT1G48140
|
DPMS3, dolichol phosphate mannose synthase 3 |
0.612 |
0.007 |
| 26 |
264399_at |
AT1G61780
|
[postsynaptic protein-related] |
0.610 |
0.055 |
| 27 |
256256_at |
AT3G11230
|
[Yippee family putative zinc-binding protein] |
0.605 |
0.013 |
| 28 |
257074_at |
AT3G19660
|
unknown |
0.603 |
0.007 |
| 29 |
250301_at |
AT5G11970
|
unknown |
0.594 |
0.068 |
| 30 |
257054_at |
AT3G15353
|
ATMT3, MT3, metallothionein 3 |
0.589 |
-0.068 |
| 31 |
266167_at |
AT2G38860
|
DJ-1e, DJ1E, YLS5 |
0.588 |
0.217 |
| 32 |
264635_at |
AT1G65500
|
unknown |
0.588 |
0.218 |
| 33 |
253990_at |
AT4G26160
|
ACHT1, atypical CYS HIS rich thioredoxin 1 |
0.586 |
-0.028 |
| 34 |
256301_at |
AT1G69510
|
[cAMP-regulated phosphoprotein 19-related protein] |
0.582 |
-0.080 |
| 35 |
245164_at |
AT2G33210
|
HSP60-2, heat shock protein 60-2 |
0.573 |
0.166 |
| 36 |
249380_at |
AT5G40370
|
GRXC2, glutaredoxin C2 |
0.571 |
0.037 |
| 37 |
264362_at |
AT1G03290
|
unknown |
0.568 |
0.164 |
| 38 |
264938_at |
AT1G60610
|
[SBP (S-ribonuclease binding protein) family protein] |
0.557 |
-0.048 |
| 39 |
254577_at |
AT4G19450
|
[Major facilitator superfamily protein] |
0.555 |
-0.020 |
| 40 |
266476_at |
AT2G31090
|
unknown |
0.543 |
0.045 |
| 41 |
264657_at |
AT1G09100
|
RPT5B, 26S proteasome AAA-ATPase subunit RPT5B |
0.519 |
0.144 |
| 42 |
265456_at |
AT2G46505
|
SDH4, succinate dehydrogenase subunit 4 |
0.503 |
0.043 |
| 43 |
253609_at |
AT4G30190
|
AHA2, H(+)-ATPase 2, HA2, H(+)-ATPase 2, PMA2, PLASMA MEMBRANE PROTON ATPASE 2 |
0.501 |
-0.023 |
| 44 |
267567_at |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
0.497 |
0.498 |
| 45 |
262837_at |
AT1G14830
|
ADL1C, DYNAMIN-like 1C, ADL5, ARABIDOPSIS DYNAMIN-LIKE PROTEIN 5, DL1C, DYNAMIN-like 1C, DRP1C, DYNAMIN RELATED PROTEIN 1C |
0.494 |
-0.013 |
| 46 |
254660_at |
AT4G18250
|
[receptor serine/threonine kinase, putative] |
0.493 |
0.340 |
| 47 |
266317_at |
AT2G27030
|
ACAM-2, CAM5, calmodulin 5 |
0.490 |
0.015 |
| 48 |
252549_at |
AT3G45860
|
CRK4, cysteine-rich RLK (RECEPTOR-like protein kinase) 4 |
0.490 |
0.644 |
| 49 |
260579_at |
AT2G47380
|
[Cytochrome c oxidase subunit Vc family protein] |
0.489 |
0.047 |
| 50 |
255816_at |
AT2G33470
|
ATGLTP1, ARABIDOPSIS GLYCOLIPID TRANSFER PROTEIN 1, GLTP1, glycolipid transfer protein 1 |
0.488 |
0.011 |
| 51 |
261486_at |
AT1G14510
|
AL7, alfin-like 7, AtAL7 |
0.486 |
0.031 |
| 52 |
253274_at |
AT4G34200
|
EDA9, embryo sac development arrest 9, PGDH1, phosphoglycerate dehydrogenase 1 |
0.485 |
0.082 |
| 53 |
248126_at |
AT5G54760
|
[Translation initiation factor SUI1 family protein] |
0.481 |
0.009 |
| 54 |
263631_at |
AT2G04900
|
unknown |
0.478 |
0.044 |
| 55 |
259008_at |
AT3G09390
|
ATMT-1, ARABIDOPSIS THALIANA METALLOTHIONEIN-1, ATMT-K, ARABIDOPSIS THALIANA METALLOTHIONEIN-K, MT2A, metallothionein 2A |
0.478 |
-0.003 |
| 56 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
0.477 |
0.178 |
| 57 |
247532_at |
AT5G61560
|
[U-box domain-containing protein kinase family protein] |
0.477 |
0.102 |
| 58 |
261955_at |
AT1G64520
|
RPN12a, regulatory particle non-ATPase 12A |
0.468 |
0.099 |
| 59 |
248513_at |
AT5G50500
|
unknown |
0.467 |
-0.017 |
| 60 |
250006_at |
AT5G18660
|
PCB2, PALE-GREEN AND CHLOROPHYLL B REDUCED 2 |
-0.888 |
-0.055 |
| 61 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.861 |
-0.480 |
| 62 |
247593_at |
AT5G60790
|
ABCF1, ATP-binding cassette F1, ATGCN1, ARABIDOPSIS THALIANA GENERAL CONTROL NON-REPRESSIBLE 1, GCN1, GENERAL CONTROL NON-REPRESSIBLE 1 |
-0.673 |
0.042 |
| 63 |
254085_at |
AT4G24960
|
ATHVA22D, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE D, HVA22D, HVA22 homologue D |
-0.664 |
-0.169 |
| 64 |
254813_at |
AT4G12350
|
AtMYB42, myb domain protein 42, MYB42, myb domain protein 42 |
-0.642 |
-0.015 |
| 65 |
254667_at |
AT4G18280
|
[glycine-rich cell wall protein-related] |
-0.615 |
-0.136 |
| 66 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
-0.601 |
0.059 |
| 67 |
254772_at |
AT4G13390
|
EXT12, extensin 12 |
-0.511 |
-0.026 |
| 68 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
-0.498 |
-0.381 |
| 69 |
253684_at |
AT4G29690
|
[Alkaline-phosphatase-like family protein] |
-0.491 |
0.014 |
| 70 |
258893_at |
AT3G05660
|
AtRLP33, receptor like protein 33, RLP33, receptor like protein 33 |
-0.485 |
-0.084 |
| 71 |
247439_at |
AT5G62670
|
AHA11, H(+)-ATPase 11, HA11, H(+)-ATPase 11 |
-0.482 |
-0.254 |
| 72 |
264371_at |
AT1G12090
|
ELP, extensin-like protein |
-0.476 |
-0.110 |
| 73 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-0.468 |
-0.334 |
| 74 |
267631_at |
AT2G42150
|
[DNA-binding bromodomain-containing protein] |
-0.467 |
0.071 |
| 75 |
258181_at |
AT3G21670
|
AtNPF6.4, NPF6.4, NRT1/ PTR family 6.4 |
-0.456 |
-0.122 |
| 76 |
253197_at |
AT4G35250
|
HCF244, high chlorophyll fluorescence phenotype 244 |
-0.450 |
0.027 |
| 77 |
259431_at |
AT1G01620
|
PIP1;3, PLASMA MEMBRANE INTRINSIC PROTEIN 1;3, PIP1C, plasma membrane intrinsic protein 1C, TMP-B |
-0.434 |
-0.160 |
| 78 |
260982_at |
AT1G53520
|
AtFAP3, FAP3, fatty-acid-binding protein 3 |
-0.428 |
-0.260 |
| 79 |
251996_at |
AT3G52840
|
BGAL2, beta-galactosidase 2 |
-0.401 |
-0.138 |
| 80 |
262897_at |
AT1G59840
|
CCB4, cofactor assembly of complex C |
-0.395 |
-0.082 |
| 81 |
245743_at |
AT1G51080
|
unknown |
-0.383 |
-0.061 |
| 82 |
249522_at |
AT5G38700
|
unknown |
-0.382 |
0.005 |
| 83 |
260236_at |
AT1G74470
|
[Pyridine nucleotide-disulphide oxidoreductase family protein] |
-0.373 |
-0.004 |
| 84 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.372 |
-0.481 |
| 85 |
251227_at |
AT3G62700
|
ABCC14, ATP-binding cassette C14, ATMRP10, multidrug resistance-associated protein 10, MRP10, multidrug resistance-associated protein 10 |
-0.370 |
-0.045 |
| 86 |
263678_at |
AT1G04420
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.365 |
-0.055 |
| 87 |
258143_at |
AT3G18170
|
[Glycosyltransferase family 61 protein] |
-0.359 |
0.003 |
| 88 |
253971_at |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
-0.355 |
-0.397 |
| 89 |
246919_at |
AT5G25460
|
DGR2, DUF642 L-GalL responsive gene 2 |
-0.352 |
-0.221 |
| 90 |
253835_at |
AT4G27820
|
BGLU9, beta glucosidase 9 |
-0.348 |
0.080 |
| 91 |
264769_at |
AT1G61350
|
[ARM repeat superfamily protein] |
-0.347 |
-0.065 |
| 92 |
249766_at |
AT5G24070
|
[Peroxidase superfamily protein] |
-0.345 |
-0.013 |
| 93 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.333 |
-0.077 |
| 94 |
259578_at |
AT1G27990
|
unknown |
-0.321 |
-0.011 |
| 95 |
247845_at |
AT5G58090
|
[O-Glycosyl hydrolases family 17 protein] |
-0.321 |
-0.034 |
| 96 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
-0.316 |
-0.086 |
| 97 |
250963_at |
AT5G03000
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.314 |
0.028 |
| 98 |
247376_at |
AT5G63310
|
ATNDPK2, ARABIDOPSIS NUCLEOSIDE DIPHOSPHATE KINASE 2, NDPK IA, NUCLEOSIDE DIPHOSPHATE KINASE IA, NDPK IA IA, NDPK1A, NDP KINASE 1A, NDPK2, nucleoside diphosphate kinase 2 |
-0.313 |
-0.020 |
| 99 |
265351_at |
AT2G22630
|
AGL17, AGAMOUS-like 17 |
-0.313 |
0.010 |
| 100 |
254274_at |
AT4G22770
|
AHL2, AT-hook motif nuclear localized protein 2 |
-0.308 |
-0.129 |
| 101 |
252391_at |
AT3G47860
|
CHL, chloroplastic lipocalin |
-0.306 |
-0.173 |
| 102 |
252215_at |
AT3G50240
|
KICP-02 |
-0.305 |
-0.142 |
| 103 |
257003_at |
AT3G14110
|
FLU, FLUORESCENT IN BLUE LIGHT |
-0.299 |
-0.076 |
| 104 |
262569_at |
AT1G15180
|
[MATE efflux family protein] |
-0.299 |
-0.131 |
| 105 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
-0.295 |
0.242 |
| 106 |
267635_at |
AT2G42220
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.294 |
-0.068 |
| 107 |
249875_at |
AT5G23120
|
HCF136, HIGH CHLOROPHYLL FLUORESCENCE 136 |
-0.294 |
-0.032 |
| 108 |
265768_at |
AT2G48020
|
[Major facilitator superfamily protein] |
-0.294 |
0.035 |
| 109 |
247028_at |
AT5G67100
|
ICU2, INCURVATA2 |
-0.293 |
-0.010 |
| 110 |
251193_at |
AT3G62910
|
APG3, ALBINO AND PALE GREEN, AtcpRF1, Arabidopsis thaliana chloroplast RF1, cpRF1, chloroplast ribosome release factor 1 |
-0.292 |
-0.055 |
| 111 |
266509_at |
AT2G47940
|
DEG2, degradation of periplasmic proteins 2, DEGP2, DEGP protease 2, EMB3117, EMBRYO DEFECTIVE 3117 |
-0.292 |
-0.112 |
| 112 |
247891_at |
AT5G57960
|
[GTP-binding protein, HflX] |
-0.291 |
-0.030 |
| 113 |
251133_at |
AT5G01240
|
LAX1, like AUXIN RESISTANT 1 |
-0.288 |
-0.206 |
| 114 |
249361_at |
AT5G40540
|
[Protein kinase superfamily protein] |
-0.285 |
0.026 |
| 115 |
262176_at |
AT1G74960
|
ATKAS2, ARABIDOPSIS BETA-KETOACYL-ACP SYNTHETASE 2, FAB1, fatty acid biosynthesis 1, KAS2, BETA-KETOACYL-ACP SYNTHETASE 2 |
-0.282 |
-0.058 |
| 116 |
246613_at |
AT5G35360
|
CAC2, acetyl Co-enzyme a carboxylase biotin carboxylase subunit |
-0.281 |
-0.076 |
| 117 |
259707_at |
AT1G77490
|
TAPX, thylakoidal ascorbate peroxidase |
-0.278 |
-0.140 |