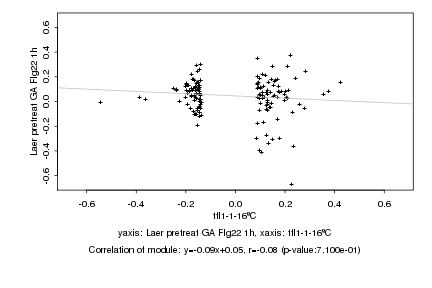
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
tfl1-1-16ºC |
Log2 signal ratio
Laer pretreat GA Flg22 1h |
| 1 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.420 |
0.157 |
| 2 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
0.375 |
0.085 |
| 3 |
267509_at |
AT2G45660
|
AGL20, AGAMOUS-like 20, ATSOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1, SOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1 |
0.352 |
0.060 |
| 4 |
259352_at |
AT3G05170
|
[Phosphoglycerate mutase family protein] |
0.280 |
0.249 |
| 5 |
253859_at |
AT4G27657
|
unknown |
0.278 |
-0.050 |
| 6 |
247573_at |
AT5G61160
|
AACT1, anthocyanin 5-aromatic acyltransferase 1 |
0.257 |
-0.019 |
| 7 |
255585_at |
AT4G01550
|
ANAC069, NAC domain containing protein 69, NAC069, NAC domain containing protein 69, NTM2, NAC with Transmembrane Motif 2 |
0.239 |
0.189 |
| 8 |
265248_at |
AT2G43010
|
AtPIF4, PIF4, phytochrome interacting factor 4, SRL2 |
0.234 |
-0.360 |
| 9 |
246260_at |
AT1G31820
|
PUT1, POLYAMINE UPTAKE TRANSPORTER 1 |
0.230 |
-0.084 |
| 10 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
0.223 |
-0.664 |
| 11 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.218 |
0.376 |
| 12 |
246929_at |
AT5G25210
|
unknown |
0.213 |
0.090 |
| 13 |
259644_at |
AT1G68910
|
WIT2, WPP domain-interacting protein 2 |
0.208 |
0.027 |
| 14 |
266962_at |
AT2G39435
|
TRM18, TON1 Recruiting Motif 18 |
0.207 |
0.285 |
| 15 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
0.204 |
0.040 |
| 16 |
257899_at |
AT3G28345
|
ABCB15, ATP-binding cassette B15, MDR13, multi-drug resistance 13 |
0.201 |
0.083 |
| 17 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
0.196 |
0.061 |
| 18 |
258339_at |
AT3G16120
|
[Dynein light chain type 1 family protein] |
0.196 |
0.016 |
| 19 |
267391_at |
AT2G44480
|
BGLU17, beta glucosidase 17 |
0.185 |
0.085 |
| 20 |
262446_at |
AT1G49310
|
unknown |
0.176 |
0.080 |
| 21 |
262388_at |
AT1G49320
|
unknown |
0.175 |
-0.298 |
| 22 |
253672_at |
AT4G29820
|
ATCFIM-25, ARABIDOPSIS THALIANA HOMOLOG OF CFIM-25, CFIM-25, homolog of CFIM-25 |
0.172 |
0.088 |
| 23 |
255753_at |
AT1G18570
|
AtMYB51, myb domain protein 51, BW51A, BW51B, HIG1, HIGH INDOLIC GLUCOSINOLATE 1, MYB51, myb domain protein 51 |
0.170 |
0.122 |
| 24 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
0.167 |
0.034 |
| 25 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
0.167 |
0.180 |
| 26 |
250365_at |
AT5G11410
|
[Protein kinase superfamily protein] |
0.166 |
-0.139 |
| 27 |
259520_at |
AT1G12320
|
unknown |
0.161 |
0.175 |
| 28 |
263465_at |
AT2G31940
|
unknown |
0.157 |
0.054 |
| 29 |
267254_at |
AT2G23030
|
SNRK2-9, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-9, SNRK2.9, SNF1-related protein kinase 2.9 |
0.155 |
0.163 |
| 30 |
249928_at |
AT5G22250
|
AtCAF1b, CCR4- associated factor 1b, CAF1b, CCR4- associated factor 1b |
0.153 |
0.047 |
| 31 |
249309_at |
AT5G41410
|
BEL1, BELL 1 |
0.152 |
0.133 |
| 32 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
0.149 |
0.289 |
| 33 |
249807_at |
AT5G23870
|
[Pectinacetylesterase family protein] |
0.149 |
-0.307 |
| 34 |
255931_at |
AT1G12710
|
AtPP2-A12, phloem protein 2-A12, PP2-A12, phloem protein 2-A12 |
0.143 |
0.180 |
| 35 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
0.142 |
-0.015 |
| 36 |
260546_at |
AT2G43520
|
ATTI2, trypsin inhibitor protein 2, TI2, trypsin inhibitor protein 2 |
0.141 |
-0.042 |
| 37 |
248911_at |
AT5G45830
|
ATDOG1, DOG1, DELAY OF GERMINATION 1, GAAS5, germination ability after storage 5, GSQ5, GLUCOSE SENSING QTL 5 |
0.139 |
0.077 |
| 38 |
252533_at |
AT3G46110
|
[LOCATED IN: plasma membrane] |
0.137 |
-0.047 |
| 39 |
260640_at |
AT1G53350
|
[Disease resistance protein (CC-NBS-LRR class) family] |
0.132 |
0.159 |
| 40 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
0.130 |
-0.338 |
| 41 |
245543_at |
AT4G15260
|
[UDP-Glycosyltransferase superfamily protein] |
0.129 |
0.064 |
| 42 |
259753_at |
AT1G71050
|
HIPP20, heavy metal associated isoprenylated plant protein 20 |
0.126 |
-0.023 |
| 43 |
252427_at |
AT3G47640
|
PYE, POPEYE |
0.126 |
-0.000 |
| 44 |
249445_at |
AT5G39380
|
[Plant calmodulin-binding protein-related] |
0.126 |
0.097 |
| 45 |
245593_at |
AT4G14550
|
IAA14, indole-3-acetic acid inducible 14, SLR, SOLITARY ROOT |
0.126 |
-0.072 |
| 46 |
260429_at |
AT1G72450
|
JAZ6, jasmonate-zim-domain protein 6, TIFY11B, TIFY DOMAIN PROTEIN 11B |
0.126 |
0.015 |
| 47 |
260156_at |
AT1G52880
|
ANAC018, Arabidopsis NAC domain containing protein 18, ATNAM, NAM, NO APICAL MERISTEM, NARS2, NAC-REGULATED SEED MORPHOLOGY 2 |
0.125 |
-0.063 |
| 48 |
266235_at |
AT2G02360
|
AtPP2-B10, phloem protein 2-B10, PP2-B10, phloem protein 2-B10 |
0.125 |
0.081 |
| 49 |
245088_at |
AT2G39850
|
[Subtilisin-like serine endopeptidase family protein] |
0.123 |
-0.272 |
| 50 |
252539_at |
AT3G45730
|
unknown |
0.122 |
-0.028 |
| 51 |
257226_at |
AT3G27880
|
unknown |
0.118 |
0.218 |
| 52 |
257678_at |
AT3G20420
|
ATRTL2, RNASEIII-LIKE 2, RTL2, RNAse THREE-like protein 2 |
0.117 |
0.038 |
| 53 |
253791_at |
AT4G28640
|
IAA11, indole-3-acetic acid inducible 11 |
0.112 |
-0.163 |
| 54 |
256922_at |
AT3G19010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.110 |
0.124 |
| 55 |
256050_at |
AT1G07000
|
ATEXO70B2, exocyst subunit exo70 family protein B2, EXO70B2, exocyst subunit exo70 family protein B2 |
0.109 |
0.223 |
| 56 |
246162_at |
AT4G36400
|
D2HGDH, D-2-hydroxyglutarate dehydrogenase |
0.108 |
0.032 |
| 57 |
246796_at |
AT5G26770
|
unknown |
0.108 |
0.075 |
| 58 |
246270_at |
AT4G36500
|
unknown |
0.106 |
0.051 |
| 59 |
259207_at |
AT3G09050
|
unknown |
0.103 |
-0.412 |
| 60 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.101 |
-0.013 |
| 61 |
261402_at |
AT1G79670
|
RFO1, RESISTANCE TO FUSARIUM OXYSPORUM 1, WAKL22 |
0.100 |
0.117 |
| 62 |
253000_at |
AT4G38360
|
LAZ1, LAZARUS 1 |
0.100 |
0.108 |
| 63 |
249551_at |
AT5G38220
|
[alpha/beta-Hydrolases superfamily protein] |
0.099 |
0.052 |
| 64 |
264899_at |
AT1G23130
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.095 |
0.030 |
| 65 |
258782_at |
AT3G11750
|
FOLB1 |
0.095 |
-0.389 |
| 66 |
255028_at |
AT4G09890
|
unknown |
0.094 |
0.190 |
| 67 |
262413_at |
AT1G34780
|
APRL4, APR-like 4, ATAPRL4, APR-like 4 |
0.094 |
0.064 |
| 68 |
246786_at |
AT5G27410
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
0.093 |
-0.071 |
| 69 |
251752_at |
AT3G55740
|
ATPROT2, PROLINE TRANSPORTER 2, PROT2, proline transporter 2 |
0.092 |
0.114 |
| 70 |
261413_at |
AT1G07630
|
PLL5, pol-like 5 |
0.089 |
0.159 |
| 71 |
247012_at |
AT5G67580
|
ATTBP3, ATTRB2, TELOMERE REPEAT BINDING FACTOR 2, TBP3, TELOMERE-BINDING PROTEIN 3, TRB2 |
0.088 |
0.150 |
| 72 |
249109_at |
AT5G43700
|
ATAUX2-11, AUXIN INDUCIBLE 2-11, IAA4, indole-3-acetic acid inducible 4 |
0.088 |
-0.176 |
| 73 |
248934_at |
AT5G46080
|
[Protein kinase superfamily protein] |
0.088 |
0.355 |
| 74 |
264957_at |
AT1G77000
|
ATSKP2;2, ARABIDOPSIS HOMOLOG OF HOMOLOG OF HUMAN SKP2 2, SKP2B |
0.087 |
0.141 |
| 75 |
253081_at |
AT4G36210
|
unknown |
0.086 |
0.040 |
| 76 |
254161_at |
AT4G24370
|
unknown |
0.085 |
0.206 |
| 77 |
263461_at |
AT2G31800
|
[Integrin-linked protein kinase family] |
0.085 |
0.111 |
| 78 |
258920_at |
AT3G10520
|
AHB2, haemoglobin 2, ARATH GLB2, ATGLB2, ARABIDOPSIS HEMOGLOBIN 2, GLB2, HEMOGLOBIN 2, HB2, haemoglobin 2, NSHB2, NON-SYMBIOTIC HAEMOGLOBIN 2 |
0.084 |
-0.299 |
| 79 |
266393_at |
AT2G41260
|
ATM17, M17 |
-0.545 |
0.000 |
| 80 |
261651_at |
AT1G27760
|
ATSAT32, SALT-TOLERANCE 32, SAT32, SALT-TOLERANCE 32 |
-0.387 |
0.033 |
| 81 |
262719_at |
AT1G43590
|
unknown |
-0.366 |
0.021 |
| 82 |
265117_at |
AT1G62500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.252 |
0.106 |
| 83 |
250196_at |
AT5G14580
|
[polyribonucleotide nucleotidyltransferase, putative] |
-0.241 |
0.091 |
| 84 |
265234_at |
AT2G07721
|
unknown |
-0.239 |
0.100 |
| 85 |
253942_at |
AT4G27010
|
EMB2788, EMBRYO DEFECTIVE 2788 |
-0.229 |
0.004 |
| 86 |
247962_at |
AT5G56580
|
ANQ1, ARABIDOPSIS NQK1, ATMKK6, ARABIDOPSIS THALIANA MAP KINASE KINASE 6, MKK6, MAP kinase kinase 6 |
-0.205 |
0.128 |
| 87 |
262669_at |
AT1G62850
|
[Class I peptide chain release factor] |
-0.202 |
0.036 |
| 88 |
255424_at |
AT4G03340
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
-0.201 |
0.092 |
| 89 |
248675_at |
AT5G48820
|
ICK6, inhibitor/interactor with cyclin-dependent kinase, KRP3, KIP-RELATED PROTEIN 3 |
-0.200 |
0.152 |
| 90 |
259386_at |
AT1G13400
|
JGL, JAGGED-LIKE, NUB, NUBBIN |
-0.200 |
0.143 |
| 91 |
258138_at |
AT3G24495
|
ATMSH7, ARABIDOPSIS THALIANA MUTS HOMOLOG 7, MSH6-2, MUTS HOMOLOG 6-2, MSH7, MUTS homolog 7 |
-0.197 |
0.068 |
| 92 |
265154_at |
AT1G30960
|
[GTP-binding family protein] |
-0.197 |
-0.021 |
| 93 |
267118_at |
AT2G32590
|
EMB2795, EMBRYO DEFECTIVE 2795 |
-0.197 |
0.136 |
| 94 |
256924_at |
AT3G29590
|
AT5MAT |
-0.193 |
0.132 |
| 95 |
245607_at |
AT4G14330
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.188 |
0.086 |
| 96 |
265085_at |
AT1G03780
|
AtTPX2, TPX2, targeting protein for XKLP2 |
-0.186 |
0.094 |
| 97 |
255278_at |
AT4G04940
|
[transducin family protein / WD-40 repeat family protein] |
-0.185 |
-0.056 |
| 98 |
250515_at |
AT5G09570
|
[Cox19-like CHCH family protein] |
-0.181 |
0.225 |
| 99 |
257333_at |
ATMG01360
|
COX1, cytochrome oxidase |
-0.181 |
0.046 |
| 100 |
253757_at |
AT4G28950
|
ARAC7, Arabidopsis RAC-like 7, ATRAC7, ATROP9, RAC7, ROP9, RHO-related protein from plants 9 |
-0.180 |
0.053 |
| 101 |
262278_at |
AT1G68640
|
PAN, PERIANTHIA, TGA8, TGACG sequence-specific binding protein 8 |
-0.179 |
0.107 |
| 102 |
246067_at |
AT5G19410
|
ABCG23, ATP-binding cassette G23 |
-0.177 |
0.094 |
| 103 |
258871_at |
AT3G03060
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.177 |
0.183 |
| 104 |
250529_at |
AT5G08610
|
PDE340, PIGMENT DEFECTIVE 340 |
-0.172 |
-0.077 |
| 105 |
259290_at |
AT3G11520
|
CYC2, CYCLIN 2, CYCB1;3, CYCLIN B1;3 |
-0.172 |
0.112 |
| 106 |
260192_at |
AT1G67630
|
EMB2814, EMBRYO DEFECTIVE 2814, POLA2, DNA polymerase alpha 2 |
-0.170 |
0.179 |
| 107 |
255597_at |
AT4G01730
|
[DHHC-type zinc finger family protein] |
-0.168 |
0.056 |
| 108 |
258242_at |
AT3G27640
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.167 |
0.171 |
| 109 |
254537_at |
AT4G19730
|
[Glycosyl hydrolase superfamily protein] |
-0.167 |
0.009 |
| 110 |
266687_at |
AT2G19670
|
ATPRMT1A, ARABIDOPSIS THALIANA PROTEIN ARGININE METHYLTRANSFERASE 1A, PRMT1A, protein arginine methyltransferase 1A |
-0.166 |
-0.104 |
| 111 |
266280_at |
AT2G29260
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.165 |
0.093 |
| 112 |
257334_at |
ATMG01370
|
ORF111D |
-0.164 |
0.038 |
| 113 |
262752_at |
AT1G16330
|
CYCB3;1, cyclin b3;1 |
-0.164 |
0.126 |
| 114 |
260077_at |
AT1G73620
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.162 |
0.121 |
| 115 |
248036_at |
AT5G55920
|
OLI2, OLIGOCELLULA 2 |
-0.162 |
-0.103 |
| 116 |
248696_at |
AT5G48360
|
[Actin-binding FH2 (formin homology 2) family protein] |
-0.161 |
0.147 |
| 117 |
260331_at |
AT1G80270
|
PPR596, PENTATRICOPEPTIDE REPEAT 596 |
-0.159 |
-0.105 |
| 118 |
246850_at |
AT5G26860
|
LON1, lon protease 1, LON_ARA_ARA |
-0.158 |
0.028 |
| 119 |
253605_at |
AT4G30990
|
[ARM repeat superfamily protein] |
-0.158 |
0.067 |
| 120 |
263264_at |
AT2G38810
|
HTA8, histone H2A 8 |
-0.158 |
0.296 |
| 121 |
252625_at |
AT3G44750
|
ATHD2A, HD2A, HISTONE DEACETYLASE 2A, HDA3, histone deacetylase 3, HDT1 |
-0.157 |
-0.052 |
| 122 |
254862_at |
AT4G12030
|
BASS5, BILE ACID:SODIUM SYMPORTER FAMILY PROTEIN 5, BAT5, bile acid transporter 5 |
-0.155 |
-0.070 |
| 123 |
260061_at |
AT1G73690
|
AT;CDKD;1, CYCLIN-DEPENDENT KINASE D1, CAK3AT, CDKD1;1, cyclin-dependent kinase D1;1 |
-0.154 |
0.105 |
| 124 |
263612_at |
AT2G16440
|
MCM4, MINICHROMOSOME MAINTENANCE 4 |
-0.154 |
0.245 |
| 125 |
251355_at |
AT3G61100
|
[Putative endonuclease or glycosyl hydrolase] |
-0.154 |
-0.188 |
| 126 |
255958_at |
AT1G22150
|
SULTR1;3, sulfate transporter 1;3 |
-0.153 |
0.159 |
| 127 |
252442_at |
AT3G46940
|
DUT1, DUTP-PYROPHOSPHATASE-LIKE 1 |
-0.152 |
0.099 |
| 128 |
253828_at |
AT4G27970
|
SLAH2, SLAC1 homologue 2 |
-0.151 |
0.103 |
| 129 |
252237_at |
AT3G49950
|
[GRAS family transcription factor] |
-0.151 |
0.113 |
| 130 |
261364_at |
AT1G53140
|
DRP5A, Dynamin related protein 5A |
-0.150 |
0.161 |
| 131 |
245828_at |
AT1G57820
|
ORTH2, ORTHRUS 2, VIM1, VARIANT IN METHYLATION 1 |
-0.150 |
0.137 |
| 132 |
266223_at |
AT2G28790
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.148 |
0.085 |
| 133 |
260935_at |
AT1G45110
|
[Tetrapyrrole (Corrin/Porphyrin) Methylases] |
-0.148 |
-0.033 |
| 134 |
264176_at |
AT1G02110
|
unknown |
-0.148 |
-0.120 |
| 135 |
262477_at |
AT1G11220
|
unknown |
-0.148 |
0.117 |
| 136 |
254076_at |
AT4G25340
|
ATFKBP53, FKBP53, FK506 BINDING PROTEIN 53 |
-0.148 |
0.006 |
| 137 |
256864_at |
AT3G23890
|
ATTOPII, TOPII, topoisomerase II |
-0.147 |
0.073 |
| 138 |
247575_at |
AT5G61030
|
GR-RBP3, glycine-rich RNA-binding protein 3 |
-0.147 |
-0.041 |
| 139 |
256980_at |
AT3G26932
|
DRB3, dsRNA-binding protein 3 |
-0.147 |
0.021 |
| 140 |
246246_at |
AT4G37170
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.147 |
0.071 |
| 141 |
262178_at |
AT1G77860
|
KOM, KOMPEITO |
-0.147 |
0.131 |
| 142 |
248841_at |
AT5G46740
|
UBP21, ubiquitin-specific protease 21 |
-0.146 |
0.266 |
| 143 |
250785_at |
AT5G05510
|
[Mad3/BUB1 homology region 1] |
-0.146 |
0.058 |
| 144 |
244943_at |
ATMG00070
|
NAD9, NADH dehydrogenase subunit 9 |
-0.145 |
0.071 |
| 145 |
249830_at |
AT5G23300
|
PYRD, pyrimidine d |
-0.145 |
-0.083 |
| 146 |
263712_at |
AT2G20585
|
NFD6, NUCLEAR FUSION DEFECTIVE 6 |
-0.144 |
0.022 |
| 147 |
254672_at |
AT4G18420
|
unknown |
-0.143 |
0.177 |
| 148 |
264969_at |
AT1G67320
|
EMB2813, EMBRYO DEFECTIVE 2813 |
-0.142 |
0.176 |
| 149 |
262943_at |
AT1G79470
|
[Aldolase-type TIM barrel family protein] |
-0.142 |
-0.021 |
| 150 |
265895_at |
AT2G15000
|
unknown |
-0.142 |
-0.050 |
| 151 |
262486_at |
AT1G21740
|
unknown |
-0.142 |
0.050 |
| 152 |
261080_at |
AT1G07370
|
ATPCNA1, PROLIFERATING CELLULAR NUCLEAR ANTIGEN 1, PCNA1, proliferating cellular nuclear antigen 1 |
-0.141 |
0.300 |
| 153 |
252019_at |
AT3G53040
|
[late embryogenesis abundant protein, putative / LEA protein, putative] |
-0.141 |
0.018 |
| 154 |
256125_at |
AT1G18250
|
ATLP-1 |
-0.140 |
-0.001 |
| 155 |
256797_at |
AT3G18600
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.139 |
-0.107 |