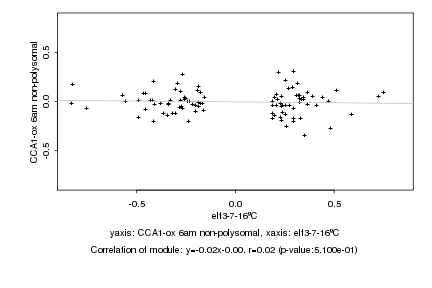
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
elf3-7-16ºC |
Log2 signal ratio
CCA1-ox 6am non-polysomal |
| 1 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
0.746 |
0.093 |
| 2 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.723 |
0.054 |
| 3 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.587 |
-0.130 |
| 4 |
248048_at |
AT5G56080
|
ATNAS2, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 2, NAS2, nicotianamine synthase 2 |
0.508 |
0.116 |
| 5 |
260030_at |
AT1G68880
|
AtbZIP, basic leucine-zipper 8, bZIP, basic leucine-zipper 8 |
0.478 |
-0.267 |
| 6 |
256779_at |
AT3G13784
|
AtcwINV5, cell wall invertase 5, CWINV5, cell wall invertase 5 |
0.467 |
0.002 |
| 7 |
245982_at |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
0.436 |
0.043 |
| 8 |
264638_at |
AT1G65480
|
FT, FLOWERING LOCUS T, RSB8, REDUCED STEM BRANCHING 8 |
0.408 |
-0.032 |
| 9 |
251293_at |
AT3G61930
|
unknown |
0.389 |
0.054 |
| 10 |
263391_at |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
0.363 |
0.092 |
| 11 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.362 |
-0.029 |
| 12 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
0.346 |
-0.344 |
| 13 |
264893_at |
AT1G23140
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.343 |
0.051 |
| 14 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.340 |
0.028 |
| 15 |
266778_at |
AT2G29090
|
CYP707A2, cytochrome P450, family 707, subfamily A, polypeptide 2 |
0.334 |
0.023 |
| 16 |
264958_at |
AT1G76960
|
unknown |
0.329 |
-0.172 |
| 17 |
257689_at |
AT3G12820
|
AtMYB10, myb domain protein 10, MYB10, myb domain protein 10 |
0.324 |
0.040 |
| 18 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.324 |
0.065 |
| 19 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
0.320 |
-0.001 |
| 20 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
0.316 |
0.069 |
| 21 |
257793_at |
AT3G26960
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.314 |
0.193 |
| 22 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
0.307 |
0.071 |
| 23 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
0.294 |
-0.166 |
| 24 |
261684_at |
AT1G47400
|
unknown |
0.293 |
-0.201 |
| 25 |
256924_at |
AT3G29590
|
AT5MAT |
0.293 |
0.306 |
| 26 |
254371_at |
AT4G21760
|
BGLU47, beta-glucosidase 47 |
0.289 |
-0.061 |
| 27 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
0.288 |
0.146 |
| 28 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
0.269 |
-0.038 |
| 29 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
0.264 |
0.142 |
| 30 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
0.258 |
-0.254 |
| 31 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
0.251 |
-0.033 |
| 32 |
259352_at |
AT3G05170
|
[Phosphoglycerate mutase family protein] |
0.250 |
-0.129 |
| 33 |
253382_at |
AT4G33040
|
[Thioredoxin superfamily protein] |
0.249 |
0.220 |
| 34 |
262232_at |
AT1G68600
|
[Aluminium activated malate transporter family protein] |
0.236 |
-0.036 |
| 35 |
264211_at |
AT1G22770
|
FB, GI, GIGANTEA |
0.235 |
-0.109 |
| 36 |
255958_at |
AT1G22150
|
SULTR1;3, sulfate transporter 1;3 |
0.231 |
-0.185 |
| 37 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
0.229 |
0.056 |
| 38 |
245882_at |
AT5G09470
|
DIC3, dicarboxylate carrier 3 |
0.229 |
-0.049 |
| 39 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
0.228 |
-0.157 |
| 40 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
0.225 |
-0.015 |
| 41 |
256750_at |
AT3G27150
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.216 |
0.305 |
| 42 |
258618_at |
AT3G02885
|
GASA5, GAST1 protein homolog 5 |
0.208 |
0.030 |
| 43 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
0.206 |
0.078 |
| 44 |
253101_at |
AT4G37430
|
CYP81F1, CYTOCHROME P450 MONOOXYGENASE 81F1, CYP91A2, cytochrome P450, family 91, subfamily A, polypeptide 2 |
0.203 |
-0.033 |
| 45 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
0.193 |
0.048 |
| 46 |
265122_at |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
0.193 |
-0.137 |
| 47 |
260335_at |
AT1G74000
|
SS3, strictosidine synthase 3 |
0.185 |
-0.116 |
| 48 |
263318_at |
AT2G24762
|
AtGDU4, glutamine dumper 4, GDU4, glutamine dumper 4 |
0.185 |
-0.171 |
| 49 |
255294_at |
AT4G04750
|
[Major facilitator superfamily protein] |
0.184 |
0.005 |
| 50 |
256100_at |
AT1G13750
|
[Purple acid phosphatases superfamily protein] |
0.184 |
-0.036 |
| 51 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
-0.834 |
-0.011 |
| 52 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
-0.830 |
0.178 |
| 53 |
266544_at |
AT2G35300
|
AtLEA4-2, late embryogenesis abundant 4-2, LEA18, late embryogenesis abundant 18, LEA4-2, late embryogenesis abundant 4-2 |
-0.758 |
-0.061 |
| 54 |
256464_at |
AT1G32560
|
AtLEA4-1, Late Embryogenesis Abundant 4-1, LEA4-1, Late Embryogenesis Abundant 4-1 |
-0.577 |
0.066 |
| 55 |
260668_at |
AT1G19530
|
unknown |
-0.559 |
0.007 |
| 56 |
262719_at |
AT1G43590
|
unknown |
-0.495 |
-0.156 |
| 57 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
-0.495 |
0.013 |
| 58 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.469 |
0.085 |
| 59 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
-0.457 |
-0.072 |
| 60 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
-0.457 |
0.087 |
| 61 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
-0.431 |
0.012 |
| 62 |
266393_at |
AT2G41260
|
ATM17, M17 |
-0.421 |
0.017 |
| 63 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.420 |
0.211 |
| 64 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
-0.417 |
-0.201 |
| 65 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
-0.410 |
-0.024 |
| 66 |
264612_at |
AT1G04560
|
[AWPM-19-like family protein] |
-0.380 |
-0.014 |
| 67 |
260431_at |
AT1G68190
|
BBX27, B-box domain protein 27 |
-0.365 |
-0.118 |
| 68 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
-0.348 |
-0.140 |
| 69 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
-0.344 |
-0.013 |
| 70 |
247754_at |
AT5G59080
|
unknown |
-0.344 |
-0.026 |
| 71 |
251869_at |
AT3G54500
|
LNK2, night light-inducible and clock-regulated 2 |
-0.331 |
0.019 |
| 72 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.323 |
-0.116 |
| 73 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
-0.305 |
0.123 |
| 74 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
-0.305 |
-0.117 |
| 75 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
-0.295 |
0.187 |
| 76 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.287 |
-0.051 |
| 77 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
-0.281 |
0.108 |
| 78 |
248969_at |
AT5G45310
|
unknown |
-0.280 |
0.019 |
| 79 |
263953_at |
AT2G36050
|
ATOFP15, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 15, OFP15, ovate family protein 15 |
-0.275 |
-0.043 |
| 80 |
261026_at |
AT1G01240
|
unknown |
-0.270 |
-0.063 |
| 81 |
264379_at |
AT2G25200
|
unknown |
-0.270 |
0.277 |
| 82 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
-0.263 |
0.048 |
| 83 |
258939_at |
AT3G10020
|
unknown |
-0.262 |
0.027 |
| 84 |
247223_at |
AT5G65070
|
AGL69, AGAMOUS-like 69, FCL4, MAF4, MADS AFFECTING FLOWERING 4 |
-0.256 |
0.040 |
| 85 |
267538_at |
AT2G41870
|
[Remorin family protein] |
-0.245 |
0.005 |
| 86 |
265724_at |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
-0.242 |
-0.201 |
| 87 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
-0.234 |
0.006 |
| 88 |
256980_at |
AT3G26932
|
DRB3, dsRNA-binding protein 3 |
-0.220 |
-0.023 |
| 89 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
-0.207 |
-0.100 |
| 90 |
256024_at |
AT1G58340
|
BCD1, BUSH-AND-CHLOROTIC-DWARF 1, ZF14, ZRZ, ZRIZI |
-0.205 |
-0.036 |
| 91 |
246962_s_at |
AT5G24800
|
ATBZIP9, ARABIDOPSIS THALIANA BASIC LEUCINE ZIPPER 9, BZIP9, basic leucine zipper 9, BZO2H2, BASIC LEUCINE ZIPPER O2 HOMOLOG 2 |
-0.197 |
0.117 |
| 92 |
252765_at |
AT3G42800
|
unknown |
-0.192 |
0.159 |
| 93 |
248050_at |
AT5G56100
|
[glycine-rich protein / oleosin] |
-0.192 |
-0.041 |
| 94 |
254954_at |
AT4G10910
|
unknown |
-0.188 |
-0.003 |
| 95 |
258262_at |
AT3G15770
|
unknown |
-0.182 |
0.101 |
| 96 |
251580_at |
AT3G58450
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.180 |
-0.016 |
| 97 |
257113_at |
AT3G20130
|
CYP705A22, cytochrome P450, family 705, subfamily A, polypeptide 22, GPS1, gravity persistence signal 1 |
-0.171 |
-0.018 |
| 98 |
263128_at |
AT1G78600
|
BBX22, B-box domain protein 22, DBB3, DOUBLE B-BOX 3, LZF1, light-regulated zinc finger protein 1, STH3, SALT TOLERANCE HOMOLOG 3 |
-0.162 |
-0.085 |
| 99 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
-0.160 |
0.043 |