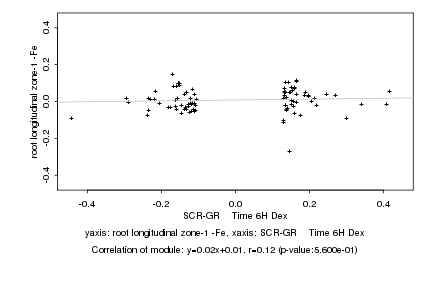
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
SCR-GR Time 6H Dex |
Log2 signal ratio
root longitudinal zone-1 -Fe |
| 1 |
260406_at |
AT1G69920
|
ATGSTU12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 12, GSTU12, glutathione S-transferase TAU 12 |
0.416 |
0.056 |
| 2 |
265405_at |
AT2G16750
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
0.406 |
-0.013 |
| 3 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
0.340 |
-0.011 |
| 4 |
264083_at |
AT2G31230
|
ATERF15, ethylene-responsive element binding factor 15, ERF15, ethylene-responsive element binding factor 15 |
0.300 |
-0.090 |
| 5 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
0.269 |
0.033 |
| 6 |
249599_at |
AT5G37990
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.245 |
0.039 |
| 7 |
259028_at |
AT3G09290
|
TAC1, telomerase activator1 |
0.219 |
-0.019 |
| 8 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
0.211 |
0.017 |
| 9 |
248389_at |
AT5G51990
|
CBF4, C-repeat-binding factor 4, DREB1D, DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN 1D |
0.203 |
0.001 |
| 10 |
254660_at |
AT4G18250
|
[receptor serine/threonine kinase, putative] |
0.197 |
0.032 |
| 11 |
266532_at |
AT2G16890
|
[UDP-Glycosyltransferase superfamily protein] |
0.197 |
0.036 |
| 12 |
266860_at |
AT2G26870
|
NPC2, non-specific phospholipase C2 |
0.188 |
0.053 |
| 13 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
0.186 |
0.035 |
| 14 |
257467_at |
AT1G31320
|
LBD4, LOB domain-containing protein 4 |
0.174 |
-0.071 |
| 15 |
264840_at |
AT1G03440
|
[Leucine-rich repeat (LRR) family protein] |
0.164 |
0.115 |
| 16 |
248276_at |
AT5G53550
|
ATYSL3, YELLOW STRIPE LIKE 3, YSL3, YELLOW STRIPE like 3 |
0.164 |
-0.004 |
| 17 |
261125_at |
AT1G04990
|
[Zinc finger C-x8-C-x5-C-x3-H type family protein] |
0.164 |
0.041 |
| 18 |
253618_at |
AT4G30420
|
UMAMIT34, Usually multiple acids move in and out Transporters 34 |
0.164 |
0.110 |
| 19 |
256359_at |
AT1G66460
|
[Protein kinase superfamily protein] |
0.159 |
-0.061 |
| 20 |
253463_at |
AT4G32105
|
[Beta-1,3-N-Acetylglucosaminyltransferase family protein] |
0.159 |
0.071 |
| 21 |
255108_at |
AT4G08740
|
unknown |
0.157 |
0.077 |
| 22 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.156 |
0.005 |
| 23 |
258213_at |
AT3G17950
|
unknown |
0.155 |
-0.023 |
| 24 |
254310_at |
AT4G22430
|
[F-box family protein] |
0.152 |
0.064 |
| 25 |
253498_at |
AT4G31890
|
[ARM repeat superfamily protein] |
0.151 |
0.009 |
| 26 |
251544_at |
AT3G58790
|
GAUT15, galacturonosyltransferase 15 |
0.150 |
0.078 |
| 27 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.149 |
-0.016 |
| 28 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
0.147 |
0.049 |
| 29 |
254178_at |
AT4G23880
|
unknown |
0.146 |
0.053 |
| 30 |
262557_at |
AT1G31330
|
PSAF, photosystem I subunit F |
0.144 |
-0.270 |
| 31 |
256989_at |
AT3G28580
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.142 |
0.106 |
| 32 |
245534_at |
AT4G15150
|
[glycine-rich protein] |
0.140 |
-0.033 |
| 33 |
258265_at |
AT3G15800
|
[Glycosyl hydrolase superfamily protein] |
0.137 |
0.027 |
| 34 |
259981_at |
AT1G76450
|
[Photosystem II reaction center PsbP family protein] |
0.137 |
-0.048 |
| 35 |
264074_at |
AT2G10780
|
[gypsy-like retrotransposon family, has a 0. P-value blast match to GB:AAD27547 polyprotein (Gypsy_Ty3-element) (Oryza sativa subsp. indica)] |
0.136 |
-0.039 |
| 36 |
262464_at |
AT1G50280
|
[Phototropic-responsive NPH3 family protein] |
0.135 |
-0.017 |
| 37 |
247180_at |
AT5G65350
|
HTR11, histone 3 11 |
0.134 |
-0.017 |
| 38 |
255916_at |
AT5G28550
|
unknown |
0.134 |
0.051 |
| 39 |
261162_at |
AT1G34440
|
unknown |
0.133 |
0.104 |
| 40 |
266564_at |
AT2G24000
|
scpl22, serine carboxypeptidase-like 22 |
0.131 |
0.033 |
| 41 |
253166_at |
AT4G35290
|
ATGLR3.2, ATGLUR2, GLR3.2, GLUTAMATE RECEPTOR 3.2, GLUR2, glutamate receptor 2 |
0.131 |
0.073 |
| 42 |
267634_at |
AT2G42100
|
[Actin-like ATPase superfamily protein] |
0.131 |
0.058 |
| 43 |
253641_at |
AT4G29980
|
unknown |
0.130 |
0.051 |
| 44 |
248359_at |
AT5G52410
|
unknown |
0.129 |
-0.110 |
| 45 |
257909_at |
AT3G25480
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
0.129 |
-0.098 |
| 46 |
258769_at |
AT3G10870
|
ATMES17, ARABIDOPSIS THALIANA METHYL ESTERASE 17, MES17, methyl esterase 17 |
0.128 |
0.021 |
| 47 |
252073_at |
AT3G51750
|
unknown |
-0.445 |
-0.089 |
| 48 |
264939_at |
AT1G60630
|
[Leucine-rich repeat protein kinase family protein] |
-0.297 |
0.021 |
| 49 |
254111_at |
AT4G24890
|
ATPAP24, ARABIDOPSIS THALIANA PURPLE ACID PHOSPHATASE 24, PAP24, purple acid phosphatase 24 |
-0.290 |
-0.004 |
| 50 |
264613_at |
AT1G04640
|
LIP2, lipoyltransferase 2 |
-0.239 |
-0.072 |
| 51 |
246432_at |
AT5G17490
|
AtRGL3, RGL3, RGA-like protein 3 |
-0.236 |
0.020 |
| 52 |
249793_at |
AT5G23680
|
[Sterile alpha motif (SAM) domain-containing protein] |
-0.236 |
-0.044 |
| 53 |
248673_at |
AT5G48780
|
[disease resistance protein (TIR-NBS class)] |
-0.230 |
0.012 |
| 54 |
248831_at |
AT5G47160
|
[YDG/SRA domain-containing protein] |
-0.219 |
0.013 |
| 55 |
247399_at |
AT5G62960
|
unknown |
-0.218 |
0.056 |
| 56 |
255429_at |
AT4G03410
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
-0.206 |
-0.009 |
| 57 |
249546_at |
AT5G38150
|
PMI15, plastid movement impaired 15 |
-0.183 |
-0.028 |
| 58 |
252936_at |
AT4G39160
|
[Homeodomain-like superfamily protein] |
-0.177 |
-0.030 |
| 59 |
256340_at |
AT1G72070
|
[Chaperone DnaJ-domain superfamily protein] |
-0.172 |
0.147 |
| 60 |
261095_at |
AT1G62930
|
RPF3, RNA processing factor 3 |
-0.169 |
0.084 |
| 61 |
265351_at |
AT2G22630
|
AGL17, AGAMOUS-like 17 |
-0.163 |
-0.022 |
| 62 |
257429_at |
AT2G27940
|
[RING/U-box superfamily protein] |
-0.163 |
0.006 |
| 63 |
260518_at |
AT1G51410
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.160 |
0.086 |
| 64 |
255169_x_at |
AT4G07940
|
unknown |
-0.160 |
-0.040 |
| 65 |
245068_at |
AT2G23260
|
UGT84B1, UDP-glucosyl transferase 84B1 |
-0.158 |
0.021 |
| 66 |
255579_at |
AT4G01460
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.155 |
0.103 |
| 67 |
255780_at |
AT1G18580
|
GAUT11, galacturonosyltransferase 11 |
-0.154 |
0.100 |
| 68 |
253822_at |
AT4G28410
|
RSA1, Root System Architecture 1 |
-0.153 |
0.101 |
| 69 |
251402_at |
AT3G60290
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.152 |
0.087 |
| 70 |
249865_at |
AT5G22820
|
[ARM repeat superfamily protein] |
-0.148 |
-0.062 |
| 71 |
262622_at |
AT1G06510
|
unknown |
-0.147 |
-0.019 |
| 72 |
263822_at |
AT2G40240
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.139 |
-0.041 |
| 73 |
261467_at |
AT1G28520
|
ATVOZ1, VASCULAR PLANT ONE ZINC FINGER PROTEIN, VOZ1, vascular plant one zinc finger protein |
-0.139 |
0.039 |
| 74 |
263586_at |
AT2G25350
|
[Phox (PX) domain-containing protein] |
-0.135 |
-0.029 |
| 75 |
266962_at |
AT2G39435
|
TRM18, TON1 Recruiting Motif 18 |
-0.133 |
0.049 |
| 76 |
256655_at |
AT3G18890
|
AtTic62, translocon at the inner envelope membrane of chloroplasts 62, Tic62, translocon at the inner envelope membrane of chloroplasts 62 |
-0.133 |
-0.040 |
| 77 |
261949_at |
AT1G64670
|
BDG1, BODYGUARD1, CED1, 9-CIS EPOXYCAROTENOID DIOXYGENASE DEFECTIVE 1 |
-0.128 |
-0.013 |
| 78 |
265302_at |
AT2G13970
|
[Mutator-like transposase family, has a 4.1e-39 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
-0.127 |
-0.027 |
| 79 |
264206_at |
AT1G22730
|
[MA3 domain-containing protein] |
-0.125 |
-0.058 |
| 80 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
-0.124 |
-0.007 |
| 81 |
257294_at |
AT3G15570
|
[Phototropic-responsive NPH3 family protein] |
-0.123 |
0.020 |
| 82 |
256792_at |
AT3G22150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.121 |
-0.051 |
| 83 |
261541_at |
AT1G63600
|
[Receptor-like protein kinase-related family protein] |
-0.119 |
-0.011 |
| 84 |
257314_at |
AT3G26590
|
[MATE efflux family protein] |
-0.118 |
0.069 |
| 85 |
255751_at |
AT1G31950
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.118 |
-0.010 |
| 86 |
257855_at |
AT3G13040
|
[myb-like HTH transcriptional regulator family protein] |
-0.114 |
-0.041 |
| 87 |
246903_at |
AT5G25750
|
unknown |
-0.113 |
0.043 |
| 88 |
255980_at |
AT1G33970
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.112 |
-0.008 |
| 89 |
252019_at |
AT3G53040
|
[late embryogenesis abundant protein, putative / LEA protein, putative] |
-0.111 |
-0.053 |
| 90 |
261838_at |
AT1G16030
|
Hsp70b, heat shock protein 70B |
-0.110 |
-0.047 |
| 91 |
247079_at |
AT5G66055
|
AKRP, ankyrin repeat protein, EMB16, EMBRYO DEFECTIVE 16, EMB2036, EMBRYO DEFECTIVE 2036 |
-0.108 |
-0.017 |
| 92 |
248483_at |
AT5G50990
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.107 |
0.012 |