|
probeID |
AGICode |
Annotation |
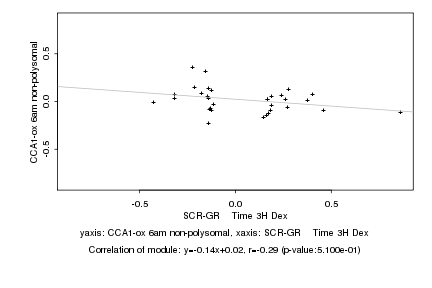
Log2 signal ratio
SCR-GR Time 3H Dex |
Log2 signal ratio
CCA1-ox 6am non-polysomal |
| 1 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
0.859 |
-0.109 |
| 2 |
248741_at |
AT5G48170
|
SLY2, SLEEPY2, SNE, SNEEZY |
0.456 |
-0.088 |
| 3 |
262325_at |
AT1G64160
|
AtDIR5, DIR5, dirigent protein 5 |
0.399 |
0.081 |
| 4 |
255422_at |
AT4G03270
|
CYCD6;1, Cyclin D6;1 |
0.375 |
0.016 |
| 5 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
0.276 |
0.135 |
| 6 |
246418_at |
AT5G16960
|
[Zinc-binding dehydrogenase family protein] |
0.268 |
-0.059 |
| 7 |
266860_at |
AT2G26870
|
NPC2, non-specific phospholipase C2 |
0.260 |
0.029 |
| 8 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.239 |
0.073 |
| 9 |
261545_at |
AT1G63530
|
unknown |
0.185 |
-0.032 |
| 10 |
265983_at |
AT2G18550
|
ATHB21, homeobox protein 21, HB-2, homeobox-2, HB21, homeobox protein 21 |
0.184 |
0.057 |
| 11 |
260023_at |
AT1G30040
|
ATGA2OX2, gibberellin 2-oxidase, GA2OX2, gibberellin 2-oxidase, GA2OX2, GIBBERELLIN 2-OXIDASE 2 |
0.181 |
-0.092 |
| 12 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
0.170 |
-0.116 |
| 13 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.167 |
0.023 |
| 14 |
249255_at |
AT5G41610
|
ATCHX18, ARABIDOPSIS THALIANA CATION/H+ EXCHANGER 18, CHX18, cation/H+ exchanger 18 |
0.165 |
0.029 |
| 15 |
259520_at |
AT1G12320
|
unknown |
0.157 |
-0.140 |
| 16 |
261222_at |
AT1G20120
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.143 |
-0.161 |
| 17 |
255814_at |
AT1G19900
|
[glyoxal oxidase-related protein] |
-0.429 |
0.000 |
| 18 |
266008_at |
AT2G37390
|
NAKR2, SODIUM POTASSIUM ROOT DEFECTIVE 2 |
-0.320 |
0.036 |
| 19 |
260542_at |
AT2G43560
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.318 |
0.079 |
| 20 |
261095_at |
AT1G62930
|
RPF3, RNA processing factor 3 |
-0.226 |
0.363 |
| 21 |
255169_x_at |
AT4G07940
|
unknown |
-0.218 |
0.148 |
| 22 |
263409_at |
AT2G04063
|
[glycine-rich protein] |
-0.179 |
0.085 |
| 23 |
264598_at |
AT1G04610
|
YUC3, YUCCA 3 |
-0.157 |
0.323 |
| 24 |
260081_at |
AT1G78170
|
unknown |
-0.146 |
0.061 |
| 25 |
245802_at |
AT1G46840
|
[F-box family protein] |
-0.145 |
0.144 |
| 26 |
265178_at |
AT1G23540
|
AtPERK12, proline-rich extensin like receptor kinase, IGI1, INFLORESCENCE GROWTH INHIBITOR 1, PERK12, proline-rich extensin-like receptor kinase 12 |
-0.142 |
0.041 |
| 27 |
262857_at |
AT1G14930
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.142 |
-0.221 |
| 28 |
263769_at |
AT2G06390
|
transposable element gene |
-0.140 |
-0.081 |
| 29 |
247596_at |
AT5G60840
|
unknown |
-0.131 |
-0.068 |
| 30 |
248734_at |
AT5G48090
|
ELP1, EDM2-like protein1 |
-0.130 |
-0.093 |
| 31 |
260059_at |
AT1G78090
|
ATTPPB, Arabidopsis thaliana trehalose-6-phosphate phosphatase B, TPPB, trehalose-6-phosphate phosphatase B |
-0.130 |
0.124 |
| 32 |
264206_at |
AT1G22730
|
[MA3 domain-containing protein] |
-0.115 |
-0.029 |