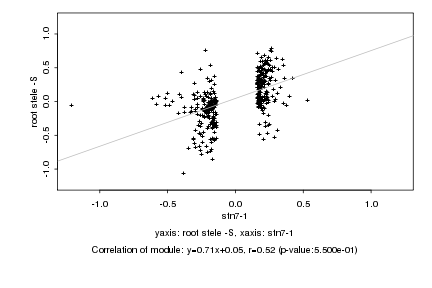
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
stn7-1 |
Log2 signal ratio
root stele -S |
| 1 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
0.529 |
0.027 |
| 2 |
259172_at |
AT3G03630
|
CS26, cysteine synthase 26 |
0.420 |
0.346 |
| 3 |
247406_at |
AT5G62920
|
ARR6, response regulator 6 |
0.396 |
0.077 |
| 4 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
0.375 |
-0.053 |
| 5 |
251935_at |
AT3G54090
|
FLN1, fructokinase-like 1 |
0.360 |
0.355 |
| 6 |
263599_at |
AT2G01830
|
AHK4, ARABIDOPSIS HISTIDINE KINASE 4, ATCRE1, CRE1, CYTOKININ RESPONSE 1, WOL, WOODEN LEG, WOL1, WOODEN LEG 1 |
0.353 |
-0.029 |
| 7 |
260343_at |
AT1G69200
|
FLN2, fructokinase-like 2 |
0.350 |
0.545 |
| 8 |
250529_at |
AT5G08610
|
PDE340, PIGMENT DEFECTIVE 340 |
0.342 |
0.636 |
| 9 |
254011_at |
AT4G26370
|
[antitermination NusB domain-containing protein] |
0.327 |
0.214 |
| 10 |
258034_at |
AT3G21300
|
[RNA methyltransferase family protein] |
0.312 |
0.483 |
| 11 |
256537_at |
AT1G33340
|
[ENTH/ANTH/VHS superfamily protein] |
0.310 |
-0.417 |
| 12 |
248709_at |
AT5G48470
|
PRDA1, PEP-Related Development Arrested 1 |
0.310 |
0.130 |
| 13 |
258545_at |
AT3G07050
|
NSN1, nucleostemin-like 1 |
0.302 |
0.638 |
| 14 |
263864_at |
AT2G04530
|
CPZ, TRZ2, TRNASE Z 2 |
0.294 |
0.325 |
| 15 |
253579_at |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
0.289 |
0.030 |
| 16 |
253777_at |
AT4G28450
|
[nucleotide binding] |
0.284 |
0.518 |
| 17 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
0.284 |
-0.524 |
| 18 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
0.283 |
0.001 |
| 19 |
250561_at |
AT5G08030
|
AtGDPD6, GDPD6, glycerophosphodiester phosphodiesterase 6 |
0.278 |
0.185 |
| 20 |
249354_at |
AT5G40480
|
EMB3012, embryo defective 3012 |
0.270 |
0.457 |
| 21 |
267166_at |
AT2G37720
|
TBL15, TRICHOME BIREFRINGENCE-LIKE 15 |
0.269 |
0.076 |
| 22 |
257207_at |
AT3G14900
|
EMB3120, EMBRYO DEFECTIVE 3120 |
0.266 |
0.504 |
| 23 |
251476_at |
AT3G59670
|
unknown |
0.265 |
0.750 |
| 24 |
256675_at |
AT3G52170
|
[DNA binding] |
0.262 |
0.798 |
| 25 |
266478_at |
AT2G31170
|
FIONA, FIONA, SYCO ARATH, cysteinyl t-RNA synthetase |
0.261 |
0.358 |
| 26 |
266957_at |
AT2G34640
|
HMR, HEMERA, PTAC12, plastid transcriptionally active 12, TAC12 |
0.256 |
0.476 |
| 27 |
250418_at |
AT5G11240
|
[transducin family protein / WD-40 repeat family protein] |
0.255 |
0.610 |
| 28 |
264357_at |
AT1G03360
|
ATRRP4, ribosomal RNA processing 4, RRP4, ribosomal RNA processing 4 |
0.255 |
0.757 |
| 29 |
256797_at |
AT3G18600
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.253 |
0.465 |
| 30 |
253949_at |
AT4G26780
|
AR192, MGE2, mitochondrial GrpE 2 |
0.252 |
0.320 |
| 31 |
263612_at |
AT2G16440
|
MCM4, MINICHROMOSOME MAINTENANCE 4 |
0.248 |
0.547 |
| 32 |
263797_at |
AT2G24570
|
ATWRKY17, WRKY17, WRKY DNA-binding protein 17 |
0.246 |
-0.332 |
| 33 |
262086_at |
AT1G56050
|
[GTP-binding protein-related] |
0.246 |
0.355 |
| 34 |
262212_at |
AT1G74890
|
ARR15, response regulator 15 |
0.243 |
0.028 |
| 35 |
257516_at |
AT1G69040
|
ACR4, ACT domain repeat 4 |
0.242 |
0.271 |
| 36 |
264471_at |
AT1G67120
|
AtMDN1, MDN1, MIDASIN 1 |
0.242 |
0.552 |
| 37 |
267562_at |
AT2G39670
|
[Radical SAM superfamily protein] |
0.241 |
0.264 |
| 38 |
263106_at |
AT2G05160
|
[CCCH-type zinc fingerfamily protein with RNA-binding domain] |
0.241 |
-0.348 |
| 39 |
254079_at |
AT4G25730
|
[FtsJ-like methyltransferase family protein] |
0.240 |
0.477 |
| 40 |
267509_at |
AT2G45660
|
AGL20, AGAMOUS-like 20, ATSOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1, SOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1 |
0.239 |
-0.008 |
| 41 |
259013_at |
AT3G07430
|
ATYLMG1-1, emb1990, embryo defective 1990, YLMG1-1 |
0.238 |
-0.197 |
| 42 |
265326_at |
AT2G18220
|
[Noc2p family] |
0.238 |
0.660 |
| 43 |
254208_at |
AT4G24175
|
unknown |
0.235 |
0.142 |
| 44 |
249977_at |
AT5G18820
|
Cpn60alpha2, chaperonin-60alpha2, EMB3007, embryo defective 3007 |
0.234 |
0.450 |
| 45 |
253285_at |
AT4G34250
|
KCS16, 3-ketoacyl-CoA synthase 16 |
0.233 |
0.069 |
| 46 |
266372_at |
AT2G41310
|
ARR8, RESPONSE REGULATOR 8, ATRR3, response regulator 3, RR3, response regulator 3 |
0.232 |
0.033 |
| 47 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
0.231 |
-0.472 |
| 48 |
257895_at |
AT3G16950
|
LPD1, lipoamide dehydrogenase 1, ptlpd1 |
0.230 |
0.609 |
| 49 |
252374_at |
AT3G48100
|
ARR5, response regulator 5, ATRR2, ARABIDOPSIS THALIANA RESPONSE REGULATOR 2, IBC6, INDUCED BY CYTOKININ 6, RR5, response regulator 5 |
0.228 |
0.085 |
| 50 |
258074_at |
AT3G25890
|
CRF11, cytokinin response factor 11 |
0.228 |
0.590 |
| 51 |
261574_at |
AT1G01190
|
CYP78A8, cytochrome P450, family 78, subfamily A, polypeptide 8 |
0.228 |
0.032 |
| 52 |
248201_at |
AT5G54180
|
PTAC15, plastid transcriptionally active 15 |
0.227 |
0.378 |
| 53 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
0.226 |
-0.022 |
| 54 |
266264_at |
AT2G27775
|
unknown |
0.225 |
0.051 |
| 55 |
266687_at |
AT2G19670
|
ATPRMT1A, ARABIDOPSIS THALIANA PROTEIN ARGININE METHYLTRANSFERASE 1A, PRMT1A, protein arginine methyltransferase 1A |
0.225 |
0.457 |
| 56 |
248075_at |
AT5G55740
|
CRR21, chlororespiratory reduction 21 |
0.223 |
0.105 |
| 57 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
0.223 |
0.151 |
| 58 |
264698_at |
AT1G70200
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.222 |
0.575 |
| 59 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
0.221 |
-0.303 |
| 60 |
245886_at |
AT5G09510
|
[Ribosomal protein S19 family protein] |
0.220 |
0.043 |
| 61 |
262750_at |
AT1G28710
|
[Nucleotide-diphospho-sugar transferase family protein] |
0.220 |
-0.360 |
| 62 |
246457_at |
AT5G16750
|
TOZ, TORMOZEMBRYO DEFECTIVE |
0.220 |
0.610 |
| 63 |
252362_at |
AT3G48500
|
PDE312, PIGMENT DEFECTIVE 312, PTAC10, PLASTID TRANSCRIPTIONALLY ACTIVE 10, TAC10 |
0.218 |
0.302 |
| 64 |
262491_at |
AT1G21650
|
SECA2 |
0.216 |
0.377 |
| 65 |
256787_at |
AT3G13790
|
ATBFRUCT1, ATCWINV1, ARABIDOPSIS THALIANA CELL WALL INVERTASE 1 |
0.216 |
0.128 |
| 66 |
263679_at |
AT1G59990
|
EMB3108, EMBRYO DEFECTIVE 3108, HS3, HEAVY SEED 3, RH22, RNA helicase 22 |
0.215 |
0.420 |
| 67 |
256978_at |
AT3G21110
|
ATPURC, PUR7, purin 7, PURC, PURIN C |
0.215 |
0.468 |
| 68 |
253295_at |
AT4G33760
|
[tRNA synthetase class II (D, K and N) family protein] |
0.214 |
0.212 |
| 69 |
259891_at |
AT1G72730
|
[DEA(D/H)-box RNA helicase family protein] |
0.214 |
0.078 |
| 70 |
258396_at |
AT3G15460
|
[Ribosomal RNA processing Brix domain protein] |
0.213 |
0.316 |
| 71 |
257891_at |
AT3G17170
|
RFC3, REGULATOR OF FATTY-ACID COMPOSITION 3 |
0.212 |
0.682 |
| 72 |
266903_at |
AT2G34570
|
MEE21, maternal effect embryo arrest 21 |
0.212 |
0.486 |
| 73 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
0.211 |
0.211 |
| 74 |
263209_at |
AT1G10522
|
PRIN2, PLASTID REDOX INSENSITIVE 2 |
0.209 |
0.265 |
| 75 |
255513_at |
AT4G02060
|
MCM7, PRL, PROLIFERA |
0.209 |
0.561 |
| 76 |
260325_at |
AT1G63940
|
MDAR6, monodehydroascorbate reductase 6 |
0.208 |
0.028 |
| 77 |
247100_at |
AT5G66520
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.208 |
0.044 |
| 78 |
247768_at |
AT5G58900
|
[Homeodomain-like transcriptional regulator] |
0.207 |
-0.096 |
| 79 |
261330_at |
AT1G44900
|
ATMCM2, MCM2, MINICHROMOSOME MAINTENANCE 2 |
0.207 |
0.568 |
| 80 |
247497_at |
AT5G61770
|
PPAN, PETER PAN-like protein |
0.206 |
0.538 |
| 81 |
256096_at |
AT1G13650
|
unknown |
0.205 |
-0.082 |
| 82 |
261296_at |
AT1G48460
|
unknown |
0.205 |
0.388 |
| 83 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.204 |
-0.560 |
| 84 |
245426_at |
AT4G17540
|
unknown |
0.204 |
0.286 |
| 85 |
260282_at |
AT1G80410
|
EMB2753, EMBRYO DEFECTIVE 2753, OMA, OMISHA |
0.203 |
0.424 |
| 86 |
259347_at |
AT3G03920
|
[H/ACA ribonucleoprotein complex, subunit Gar1/Naf1 protein] |
0.203 |
0.364 |
| 87 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
0.203 |
0.010 |
| 88 |
253252_at |
AT4G34740
|
ASE2, GLN phosphoribosyl pyrophosphate amidotransferase 2, ATASE2, GLN phosphoribosyl pyrophosphate amidotransferase 2, ATPURF2, CIA1, CHLOROPLAST IMPORT APPARATUS 1, DOV1, DIFFERENTIAL DEVELOPMENT OF VASCULAR ASSOCIATED CELLS |
0.203 |
0.418 |
| 89 |
247607_at |
AT5G60960
|
PNM1, PPR protein localized to the nucleus and mitochondria 1 |
0.202 |
0.304 |
| 90 |
257909_at |
AT3G25480
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
0.201 |
-0.177 |
| 91 |
262937_at |
AT1G79560
|
EMB1047, EMBRYO DEFECTIVE 1047, EMB156, EMBRYO DEFECTIVE 156, EMB36, EMBRYO DEFECTIVE 36, FTSH12, FTSH protease 12 |
0.199 |
0.489 |
| 92 |
249138_at |
AT5G43070
|
WPP1, WPP domain protein 1 |
0.199 |
-0.122 |
| 93 |
250837_at |
AT5G04620
|
ATBIOF, biotin F, BIO4, biotin 4, BIOF, biotin F |
0.197 |
0.423 |
| 94 |
259082_at |
AT3G04820
|
[Pseudouridine synthase family protein] |
0.195 |
0.407 |
| 95 |
262584_at |
AT1G15440
|
ATPWP2, PERIODIC TRYPTOPHAN PROTEIN 2, PWP2, periodic tryptophan protein 2 |
0.194 |
0.318 |
| 96 |
250031_at |
AT5G18240
|
ATMYR1, ARABIDOPSIS MYB-RELATED PROTEIN 1, MYR1, myb-related protein 1 |
0.194 |
0.073 |
| 97 |
246265_at |
AT1G31860
|
AT-IE, HISN2, HISTIDINE BIOSYNTHESIS 2 |
0.194 |
0.257 |
| 98 |
248891_at |
AT5G46280
|
MCM3, MINICHROMOSOME MAINTENANCE 3 |
0.194 |
0.594 |
| 99 |
258708_at |
AT3G09580
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
0.192 |
-0.098 |
| 100 |
246565_at |
AT5G15530
|
BCCP2, biotin carboxyl carrier protein 2, CAC1-B |
0.192 |
0.037 |
| 101 |
264048_at |
AT2G22400
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.191 |
0.411 |
| 102 |
250753_at |
AT5G05860
|
UGT76C2, UDP-glucosyl transferase 76C2 |
0.191 |
0.012 |
| 103 |
265895_at |
AT2G15000
|
unknown |
0.191 |
-0.049 |
| 104 |
247726_at |
AT5G59430
|
ATTRP1, TELOMERE REPEAT BINDING PROTEIN 1, TRP1, telomeric repeat binding protein 1 |
0.191 |
0.230 |
| 105 |
258884_at |
AT3G10050
|
OMR1, L-O-methylthreonine resistant 1 |
0.190 |
0.340 |
| 106 |
258225_at |
AT3G15630
|
unknown |
0.190 |
0.019 |
| 107 |
250478_at |
AT5G10250
|
DOT3, DEFECTIVELY ORGANIZED TRIBUTARIES 3 |
0.188 |
-0.076 |
| 108 |
246415_at |
AT5G17160
|
unknown |
0.188 |
0.652 |
| 109 |
263843_at |
AT2G37020
|
[Translin family protein] |
0.188 |
0.232 |
| 110 |
256862_at |
AT3G23940
|
[dehydratase family] |
0.188 |
0.290 |
| 111 |
266574_at |
AT2G23890
|
[HAD-superfamily hydrolase, subfamily IG, 5'-nucleotidase] |
0.187 |
0.225 |
| 112 |
256144_at |
AT1G48630
|
RACK1B, receptor for activated C kinase 1B, RACK1B_AT, receptor for activated C kinase 1B |
0.187 |
0.274 |
| 113 |
252410_at |
AT3G47450
|
ATNOA1, ATNOS1, NOA1, NO ASSOCIATED 1, NOS1, NITRIC OXIDE SYNTHASE 1, RIF1, RESISTANT TO INHIBITION WITH FOSMIDOMYCIN 1 |
0.186 |
0.402 |
| 114 |
258232_at |
AT3G27750
|
EMB3123, EMBRYO DEFECTIVE 3123, THA8, Thylakoid Assembly 8 |
0.186 |
-0.046 |
| 115 |
250566_at |
AT5G08070
|
TCP17, TCP domain protein 17 |
0.185 |
-0.027 |
| 116 |
258701_at |
AT3G09720
|
AtRH57, RH57, RNA helicase 57 |
0.185 |
0.474 |
| 117 |
266801_at |
AT2G22870
|
EMB2001, embryo defective 2001 |
0.185 |
0.041 |
| 118 |
259908_at |
AT1G60850
|
AAC42, ATRPAC42 |
0.183 |
0.190 |
| 119 |
264350_at |
AT1G11870
|
ATSRS, OVA7, ovule abortion 7, SRS, Seryl-tRNA synthetase |
0.183 |
0.138 |
| 120 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
0.182 |
0.084 |
| 121 |
248094_at |
AT5G55220
|
[trigger factor type chaperone family protein] |
0.181 |
0.141 |
| 122 |
253005_at |
AT4G38100
|
CURT1D, CURVATURE THYLAKOID 1D |
0.181 |
0.432 |
| 123 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
0.180 |
-0.051 |
| 124 |
253966_at |
AT4G26520
|
AtFBA7, FBA7, fructose-bisphosphate aldolase 7 |
0.180 |
0.205 |
| 125 |
260292_at |
AT1G63680
|
APG13, ALBINO OR PALE-GREEN 13, ATMURE, MURE, PDE316, PIGMENT DEFECTIVE EMBRYO 316 |
0.179 |
0.411 |
| 126 |
248295_at |
AT5G53070
|
[Ribosomal protein L9/RNase H1] |
0.179 |
0.532 |
| 127 |
246249_at |
AT4G36680
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.179 |
0.229 |
| 128 |
262094_at |
AT1G56110
|
NOP56, homolog of nucleolar protein NOP56 |
0.179 |
0.595 |
| 129 |
251538_at |
AT3G58660
|
[Ribosomal protein L1p/L10e family] |
0.178 |
0.510 |
| 130 |
266093_at |
AT2G37990
|
[ribosome biogenesis regulatory protein (RRS1) family protein] |
0.177 |
0.226 |
| 131 |
254125_at |
AT4G24670
|
TAR2, tryptophan aminotransferase related 2 |
0.176 |
-0.014 |
| 132 |
256341_at |
AT1G72040
|
AtdNK, dNK, deoxyribonucleoside kinase |
0.176 |
0.371 |
| 133 |
260596_at |
AT1G55900
|
EMB1860, embryo defective 1860, TIM50 |
0.176 |
0.218 |
| 134 |
261439_at |
AT1G28395
|
unknown |
0.175 |
0.041 |
| 135 |
259694_at |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
0.175 |
-0.479 |
| 136 |
248239_at |
AT5G53920
|
[ribosomal protein L11 methyltransferase-related] |
0.174 |
0.151 |
| 137 |
262802_at |
AT1G20930
|
CDKB2;2, cyclin-dependent kinase B2;2 |
0.173 |
0.321 |
| 138 |
245770_at |
AT1G30240
|
[FUNCTIONS IN: binding] |
0.173 |
0.471 |
| 139 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
0.173 |
-0.328 |
| 140 |
255874_at |
AT2G40550
|
ETG1, E2F target gene 1 |
0.173 |
0.334 |
| 141 |
249327_at |
AT5G40890
|
ATCLC-A, chloride channel A, ATCLCA, CLC-A, chloride channel A, CLC-A, CHLORIDE CHANNEL-A, CLCA, CHLORIDE CHANNEL A |
0.172 |
0.013 |
| 142 |
255455_at |
AT4G02930
|
[GTP binding Elongation factor Tu family protein] |
0.172 |
0.315 |
| 143 |
266035_at |
AT2G05990
|
ENR1, ENOYL-ACP REDUCTASE 1, MOD1, MOSAIC DEATH 1 |
0.172 |
0.345 |
| 144 |
248854_at |
AT5G46580
|
[pentatricopeptide (PPR) repeat-containing protein] |
0.172 |
0.048 |
| 145 |
251119_at |
AT3G63510
|
[FMN-linked oxidoreductases superfamily protein] |
0.171 |
0.019 |
| 146 |
252281_at |
AT3G49320
|
[Metal-dependent protein hydrolase] |
0.171 |
0.106 |
| 147 |
261232_at |
AT1G20220
|
[Alba DNA/RNA-binding protein] |
0.170 |
0.303 |
| 148 |
249874_at |
AT5G23070
|
AtTK1b, TK1b, thymidine kinase 1b |
0.170 |
0.334 |
| 149 |
263483_at |
AT2G04030
|
AtHsp90.5, HEAT SHOCK PROTEIN 90.5, AtHsp90C, CR88, EMB1956, EMBRYO DEFECTIVE 1956, Hsp88.1, HEAT SHOCK PROTEIN 88.1, HSP90.5, HEAT SHOCK PROTEIN 90.5 |
0.169 |
0.337 |
| 150 |
251807_at |
AT3G55400
|
OVA1, OVULE ABORTION 1 |
0.169 |
0.341 |
| 151 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
0.169 |
0.130 |
| 152 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
0.169 |
-0.083 |
| 153 |
249645_at |
AT5G36910
|
THI2.2, thionin 2.2 |
0.169 |
-0.007 |
| 154 |
251669_at |
AT3G57180
|
BPG2, BRASSINAZOLE(BRZ) INSENSITIVE PALE GREEN 2 |
0.168 |
0.499 |
| 155 |
247959_at |
AT5G57080
|
unknown |
0.168 |
-0.001 |
| 156 |
265596_at |
AT2G20020
|
ATCAF1, CAF1 |
0.168 |
0.292 |
| 157 |
256920_at |
AT3G18980
|
ETP1, EIN2 targeting protein1 |
0.167 |
0.076 |
| 158 |
258355_at |
AT3G14330
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.167 |
0.184 |
| 159 |
266889_at |
AT2G44640
|
unknown |
0.167 |
0.370 |
| 160 |
254235_at |
AT4G23750
|
CRF2, cytokinin response factor 2, TMO3, TARGET OF MONOPTEROS 3 |
0.166 |
0.377 |
| 161 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
0.166 |
-0.034 |
| 162 |
250461_at |
AT5G10010
|
HIT4, HEAT INTOLERANT 4 |
0.166 |
0.261 |
| 163 |
257856_at |
AT3G12930
|
[Lojap-related protein] |
0.166 |
-0.021 |
| 164 |
249464_at |
AT5G39710
|
EMB2745, EMBRYO DEFECTIVE 2745 |
0.165 |
0.327 |
| 165 |
247955_at |
AT5G56950
|
NAP1;3, nucleosome assembly protein 1;3, NFA03, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP A 03, NFA3, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP A3 |
0.165 |
0.027 |
| 166 |
250080_at |
AT5G16620
|
ATTIC40, translocon at the inner envelope membrane of chloroplasts 40, PDE120, pigment defective embryo 120, TIC40, translocon at the inner envelope membrane of chloroplasts 40 |
0.165 |
0.125 |
| 167 |
258316_at |
AT3G22660
|
[rRNA processing protein-related] |
0.164 |
0.302 |
| 168 |
248102_at |
AT5G55140
|
[ribosomal protein L30 family protein] |
0.164 |
-0.060 |
| 169 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.163 |
0.013 |
| 170 |
249026_at |
AT5G44785
|
OSB3, organellar single-stranded DNA binding protein 3 |
0.163 |
0.264 |
| 171 |
245085_at |
AT2G23350
|
PAB4, poly(A) binding protein 4, PABP4, POLY(A) BINDING PROTEIN 4 |
0.163 |
0.293 |
| 172 |
251371_at |
AT3G60360
|
EDA14, EMBRYO SAC DEVELOPMENT ARREST 14, UTP11, U3 SMALL NUCLEOLAR RNA-ASSOCIATED PROTEIN 11 |
0.162 |
0.279 |
| 173 |
252220_at |
AT3G49940
|
LBD38, LOB domain-containing protein 38 |
0.162 |
-0.028 |
| 174 |
254133_at |
AT4G24810
|
[Protein kinase superfamily protein] |
0.162 |
0.276 |
| 175 |
253281_at |
AT4G34138
|
UGT73B1, UDP-glucosyl transferase 73B1 |
0.162 |
0.073 |
| 176 |
248975_at |
AT5G45040
|
CYTC6A, cytochrome c6A |
0.161 |
0.325 |
| 177 |
261406_at |
AT1G18800
|
NRP2, NAP1-related protein 2 |
0.161 |
0.211 |
| 178 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
0.161 |
0.052 |
| 179 |
247378_at |
AT5G63120
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.160 |
0.292 |
| 180 |
248357_at |
AT5G52380
|
[VASCULAR-RELATED NAC-DOMAIN 6] |
0.160 |
0.450 |
| 181 |
267309_at |
AT2G19385
|
[zinc ion binding] |
0.160 |
0.375 |
| 182 |
256077_at |
AT1G18090
|
[5'-3' exonuclease family protein] |
0.160 |
0.323 |
| 183 |
250403_at |
AT5G10920
|
[L-Aspartase-like family protein] |
0.160 |
0.107 |
| 184 |
246303_at |
AT3G51870
|
[Mitochondrial substrate carrier family protein] |
0.160 |
0.220 |
| 185 |
246420_at |
AT5G16870
|
[Peptidyl-tRNA hydrolase II (PTH2) family protein] |
0.160 |
0.275 |
| 186 |
254452_at |
AT4G21100
|
DDB1B, damaged DNA binding protein 1B |
0.160 |
0.517 |
| 187 |
264689_at |
AT1G09900
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.159 |
0.717 |
| 188 |
257218_at |
AT3G15000
|
MORF8, multiple organellar RNA editing factor 8, RIP1, RNA-editing factor interacting protein 1 |
0.159 |
0.266 |
| 189 |
256065_at |
AT1G07070
|
[Ribosomal protein L35Ae family protein] |
0.159 |
0.254 |
| 190 |
260036_at |
AT1G68830
|
STN7, STT7 homolog STN7 |
-1.214 |
-0.057 |
| 191 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
-0.615 |
0.056 |
| 192 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.586 |
-0.037 |
| 193 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
-0.574 |
0.077 |
| 194 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
-0.523 |
-0.053 |
| 195 |
264958_at |
AT1G76960
|
unknown |
-0.519 |
0.057 |
| 196 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
-0.507 |
0.121 |
| 197 |
260140_at |
AT1G66390
|
ATMYB90, MYB90, myb domain protein 90, PAP2, PRODUCTION OF ANTHOCYANIN PIGMENT 2 |
-0.492 |
-0.055 |
| 198 |
259385_at |
AT1G13470
|
unknown |
-0.467 |
0.006 |
| 199 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
-0.424 |
-0.165 |
| 200 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.413 |
0.112 |
| 201 |
250344_at |
AT5G11930
|
[Thioredoxin superfamily protein] |
-0.404 |
0.063 |
| 202 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-0.398 |
0.434 |
| 203 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
-0.389 |
-1.053 |
| 204 |
252977_at |
AT4G38560
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
-0.381 |
-0.085 |
| 205 |
262092_at |
AT1G56150
|
SAUR71, SMALL AUXIN UPREGULATED 71 |
-0.380 |
-0.154 |
| 206 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
-0.350 |
-0.695 |
| 207 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
-0.334 |
-0.149 |
| 208 |
264434_at |
AT1G10340
|
[Ankyrin repeat family protein] |
-0.329 |
-0.137 |
| 209 |
262232_at |
AT1G68600
|
[Aluminium activated malate transporter family protein] |
-0.325 |
-0.079 |
| 210 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.316 |
-0.560 |
| 211 |
251928_at |
AT3G53980
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.315 |
-0.537 |
| 212 |
259766_at |
AT1G64360
|
unknown |
-0.311 |
0.117 |
| 213 |
256924_at |
AT3G29590
|
AT5MAT |
-0.309 |
0.041 |
| 214 |
261177_at |
AT1G04770
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.309 |
0.270 |
| 215 |
262357_at |
AT1G73040
|
[Mannose-binding lectin superfamily protein] |
-0.306 |
0.092 |
| 216 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
-0.306 |
0.043 |
| 217 |
252060_at |
AT3G52430
|
ATPAD4, ARABIDOPSIS PHYTOALEXIN DEFICIENT 4, PAD4, PHYTOALEXIN DEFICIENT 4 |
-0.304 |
-0.608 |
| 218 |
256789_at |
AT3G13672
|
SINA2 |
-0.301 |
-0.142 |
| 219 |
264893_at |
AT1G23140
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.297 |
-0.668 |
| 220 |
247071_at |
AT5G66640
|
DAR3, DA1-related protein 3 |
-0.296 |
-0.424 |
| 221 |
254178_at |
AT4G23880
|
unknown |
-0.295 |
-0.104 |
| 222 |
256569_at |
AT3G19550
|
unknown |
-0.287 |
-0.038 |
| 223 |
256509_at |
AT1G75300
|
[NmrA-like negative transcriptional regulator family protein] |
-0.286 |
-0.192 |
| 224 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.285 |
-0.088 |
| 225 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
-0.283 |
0.135 |
| 226 |
251644_at |
AT3G57540
|
[Remorin family protein] |
-0.280 |
0.054 |
| 227 |
265913_at |
AT2G25625
|
unknown |
-0.280 |
0.050 |
| 228 |
261535_at |
AT1G01725
|
unknown |
-0.274 |
-0.352 |
| 229 |
245385_at |
AT4G14020
|
[Rapid alkalinization factor (RALF) family protein] |
-0.272 |
-0.661 |
| 230 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
-0.271 |
-0.464 |
| 231 |
255957_at |
AT1G22160
|
unknown |
-0.269 |
-0.462 |
| 232 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
-0.266 |
-0.292 |
| 233 |
246403_at |
AT1G57590
|
[Pectinacetylesterase family protein] |
-0.264 |
0.476 |
| 234 |
255302_at |
AT4G04830
|
ATMSRB5, methionine sulfoxide reductase B5, MSRB5, methionine sulfoxide reductase B5 |
-0.264 |
-0.197 |
| 235 |
257074_at |
AT3G19660
|
unknown |
-0.262 |
-0.480 |
| 236 |
266761_at |
AT2G47130
|
AtSDR3, SDR3, short-chain dehydrogenase/reductase 2 |
-0.262 |
0.085 |
| 237 |
255028_at |
AT4G09890
|
unknown |
-0.261 |
-0.714 |
| 238 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.259 |
-0.358 |
| 239 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.254 |
-0.774 |
| 240 |
254500_at |
AT4G20110
|
BP80-3;1, binding protein of 80 kDa 3;1, VSR3;1, VACUOLAR SORTING RECEPTOR 3;1, VSR7, VACUOLAR SORTING RECEPTOR 7 |
-0.251 |
-0.313 |
| 241 |
266516_at |
AT2G47880
|
[Glutaredoxin family protein] |
-0.250 |
-0.109 |
| 242 |
251054_at |
AT5G01540
|
LecRK-VI.2, L-type lectin receptor kinase-VI.2, LECRKA4.1, lectin receptor kinase a4.1 |
-0.249 |
-0.195 |
| 243 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
-0.248 |
-0.514 |
| 244 |
251026_at |
AT5G02200
|
FHL, far-red-elongated hypocotyl1-like |
-0.246 |
-0.598 |
| 245 |
260943_at |
AT1G45145
|
ATH5, THIOREDOXIN H-TYPE 5, ATTRX5, thioredoxin H-type 5, LIV1, LOCUS OF INSENSITIVITY TO VICTORIN 1, TRX-h5, THIOREDOXIN H-TYPE 5, TRX5, thioredoxin H-type 5 |
-0.243 |
-0.096 |
| 246 |
248321_at |
AT5G52740
|
[Copper transport protein family] |
-0.242 |
0.041 |
| 247 |
255620_at |
AT4G01380
|
[plastocyanin-like domain-containing protein] |
-0.241 |
0.025 |
| 248 |
260840_at |
AT1G29050
|
TBL38, TRICHOME BIREFRINGENCE-LIKE 38 |
-0.241 |
-0.454 |
| 249 |
254266_at |
AT4G23130
|
CRK5, cysteine-rich RLK (RECEPTOR-like protein kinase) 5, RLK6, RECEPTOR-LIKE PROTEIN KINASE 6 |
-0.238 |
0.058 |
| 250 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
-0.237 |
0.001 |
| 251 |
254186_at |
AT4G24010
|
ATCSLG1, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G1, CSLG1, cellulose synthase like G1 |
-0.235 |
0.018 |
| 252 |
254153_at |
AT4G24450
|
ATGWD2, GWD3, PWD, phosphoglucan, water dikinase |
-0.233 |
0.146 |
| 253 |
265597_at |
AT2G20142
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.233 |
-0.208 |
| 254 |
261526_at |
AT1G14370
|
APK2A, protein kinase 2A, Kin1, kinase 1, PBL2, PBS1-like 2 |
-0.231 |
-0.035 |
| 255 |
246620_at |
AT5G36220
|
CYP81D1, cytochrome P450, family 81, subfamily D, polypeptide 1, CYP91A1, CYTOCHROME P450 91A1 |
-0.231 |
-0.119 |
| 256 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
-0.230 |
-0.230 |
| 257 |
258890_at |
AT3G05690
|
ATHAP2B, HEME ACTIVATOR PROTEIN (YEAST) HOMOLOG 2B, HAP2B, HEME ACTIVATOR PROTEIN (YEAST) HOMOLOG 2B, NF-YA2, nuclear factor Y, subunit A2, UNE8, UNFERTILIZED EMBRYO SAC 8 |
-0.229 |
-0.144 |
| 258 |
266977_at |
AT2G39420
|
[alpha/beta-Hydrolases superfamily protein] |
-0.229 |
-0.227 |
| 259 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
-0.226 |
0.118 |
| 260 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-0.225 |
0.768 |
| 261 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
-0.224 |
-0.100 |
| 262 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
-0.220 |
-0.138 |
| 263 |
260148_at |
AT1G52800
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.220 |
-0.086 |
| 264 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
-0.220 |
-0.151 |
| 265 |
251768_at |
AT3G55940
|
[Phosphoinositide-specific phospholipase C family protein] |
-0.219 |
-0.167 |
| 266 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
-0.215 |
0.082 |
| 267 |
255243_at |
AT4G05590
|
unknown |
-0.214 |
-0.656 |
| 268 |
246939_at |
AT5G25390
|
SHN3, shine3 |
-0.212 |
-0.068 |
| 269 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
-0.211 |
-0.753 |
| 270 |
259403_at |
AT1G17745
|
PGDH, 3-phosphoglycerate dehydrogenase, PGDH2, phosphoglycerate dehydrogenase 2 |
-0.210 |
-0.111 |
| 271 |
252888_at |
AT4G39210
|
APL3 |
-0.210 |
0.341 |
| 272 |
250723_at |
AT5G06300
|
LOG7, LONELY GUY 7 |
-0.210 |
0.142 |
| 273 |
266170_at |
AT2G39050
|
ArathEULS3, EULS3, Euonymus lectin S3 |
-0.207 |
-0.211 |
| 274 |
252533_at |
AT3G46110
|
[LOCATED IN: plasma membrane] |
-0.207 |
-0.395 |
| 275 |
247747_at |
AT5G59000
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-0.206 |
-0.051 |
| 276 |
253818_at |
AT4G28330
|
unknown |
-0.206 |
-0.003 |
| 277 |
250028_at |
AT5G18130
|
unknown |
-0.206 |
-0.363 |
| 278 |
247041_at |
AT5G67180
|
TOE3, target of early activation tagged (EAT) 3 |
-0.206 |
0.062 |
| 279 |
255599_at |
AT4G01010
|
ATCNGC13, CYCLIC NUCLEOTIDE-GATED CHANNEL 13, CNGC13, cyclic nucleotide-gated channel 13 |
-0.204 |
-0.211 |
| 280 |
260541_at |
AT2G43530
|
[Scorpion toxin-like knottin superfamily protein] |
-0.203 |
-0.007 |
| 281 |
264968_at |
AT1G67360
|
[Rubber elongation factor protein (REF)] |
-0.203 |
-0.121 |
| 282 |
263433_at |
AT2G22240
|
ATIPS2, INOSITOL 3-PHOSPHATE SYNTHASE 2, ATMIPS2, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 2, MIPS2, myo-inositol-1-phosphate synthase 2 |
-0.202 |
-0.197 |
| 283 |
261803_at |
AT1G30500
|
NF-YA7, nuclear factor Y, subunit A7 |
-0.201 |
-0.048 |
| 284 |
249346_at |
AT5G40780
|
LHT1, lysine histidine transporter 1 |
-0.199 |
-0.161 |
| 285 |
251722_at |
AT3G56200
|
[Transmembrane amino acid transporter family protein] |
-0.197 |
-0.086 |
| 286 |
246600_at |
AT5G14930
|
SAG101, senescence-associated gene 101 |
-0.197 |
-0.101 |
| 287 |
254343_at |
AT4G21990
|
APR3, APS reductase 3, ATAPR3, PRH-26, PRH26, PAPS REDUCTASE HOMOLOG 26 |
-0.196 |
0.298 |
| 288 |
254242_at |
AT4G23200
|
CRK12, cysteine-rich RLK (RECEPTOR-like protein kinase) 12 |
-0.195 |
-0.033 |
| 289 |
255340_at |
AT4G04490
|
CRK36, cysteine-rich RLK (RECEPTOR-like protein kinase) 36 |
-0.195 |
-0.015 |
| 290 |
266017_at |
AT2G18690
|
unknown |
-0.195 |
-0.239 |
| 291 |
253414_at |
AT4G33050
|
AtIQM1, EDA39, embryo sac development arrest 39, IQM1, IQ-motif protein 1 |
-0.195 |
-0.162 |
| 292 |
254784_at |
AT4G12720
|
AtNUDT7, Arabidopsis thaliana Nudix hydrolase homolog 7, GFG1, GROWTH FACTOR GENE 1, NUDT7 |
-0.190 |
-0.101 |
| 293 |
247577_at |
AT5G61290
|
[Flavin-binding monooxygenase family protein] |
-0.190 |
-0.551 |
| 294 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
-0.190 |
0.120 |
| 295 |
256676_at |
AT3G52180
|
ATPTPKIS1, ATSEX4, DSP4, DUAL-SPECIFICITY PROTEIN PHOSPHATASE 4, SEX4, STARCH-EXCESS 4 |
-0.190 |
0.535 |
| 296 |
253854_at |
AT4G27900
|
[CCT motif family protein] |
-0.190 |
0.011 |
| 297 |
254185_at |
AT4G23990
|
ATCSLG3, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE-LIKE G3, CSLG3, cellulose synthase like G3 |
-0.188 |
0.113 |
| 298 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
-0.187 |
-0.055 |
| 299 |
264590_at |
AT2G17710
|
unknown |
-0.186 |
-0.737 |
| 300 |
257122_at |
AT3G20250
|
APUM5, pumilio 5, PUM5, pumilio 5 |
-0.185 |
-0.067 |
| 301 |
253382_at |
AT4G33040
|
[Thioredoxin superfamily protein] |
-0.184 |
-0.327 |
| 302 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
-0.183 |
-0.300 |
| 303 |
262286_at |
AT1G68585
|
unknown |
-0.182 |
-0.006 |
| 304 |
266695_at |
AT2G19810
|
AtOZF1, AtTZF2, OZF1, Oxidation-related Zinc Finger 1, TZF2, tandem zinc finger 2 |
-0.181 |
-0.080 |
| 305 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
-0.181 |
-0.709 |
| 306 |
248072_at |
AT5G55680
|
[glycine-rich protein] |
-0.181 |
0.194 |
| 307 |
265728_at |
AT2G31990
|
[Exostosin family protein] |
-0.181 |
-0.595 |
| 308 |
258263_at |
AT3G15780
|
unknown |
-0.180 |
-0.120 |
| 309 |
264902_at |
AT1G23060
|
MDP40, MICROTUBULE DESTABILIZING PROTEIN 40 |
-0.179 |
-0.082 |
| 310 |
253317_at |
AT4G33960
|
unknown |
-0.179 |
0.026 |
| 311 |
253358_at |
AT4G32940
|
GAMMA-VPE, gamma vacuolar processing enzyme, GAMMAVPE |
-0.179 |
-0.092 |
| 312 |
247775_at |
AT5G58690
|
ATPLC5, PLC5, phosphatidylinositol-speciwc phospholipase C5 |
-0.178 |
-0.027 |
| 313 |
251591_at |
AT3G57680
|
[Peptidase S41 family protein] |
-0.178 |
0.047 |
| 314 |
263881_at |
AT2G21820
|
unknown |
-0.178 |
-0.020 |
| 315 |
254716_at |
AT4G13560
|
UNE15, unfertilized embryo sac 15 |
-0.178 |
0.056 |
| 316 |
247264_at |
AT5G64530
|
ANAC104, Arabidopsis NAC domain containing protein 104, XND1, xylem NAC domain 1 |
-0.178 |
-0.049 |
| 317 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
-0.177 |
0.320 |
| 318 |
246682_at |
AT5G33290
|
XGD1, xylogalacturonan deficient 1 |
-0.177 |
-0.090 |
| 319 |
266581_at |
AT2G46140
|
[Late embryogenesis abundant protein] |
-0.175 |
-0.101 |
| 320 |
248271_at |
AT5G53420
|
[CCT motif family protein] |
-0.175 |
-0.084 |
| 321 |
259561_at |
AT1G21250
|
AtWAK1, PRO25, WAK1, cell wall-associated kinase 1 |
-0.175 |
0.010 |
| 322 |
265957_at |
AT2G37300
|
ABCI16, ATP-binding cassette I16 |
-0.174 |
-0.388 |
| 323 |
250863_at |
AT5G04750
|
[F1F0-ATPase inhibitor protein, putative] |
-0.174 |
-0.382 |
| 324 |
258362_at |
AT3G14280
|
unknown |
-0.173 |
-0.858 |
| 325 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
-0.173 |
-0.544 |
| 326 |
251790_at |
AT3G55470
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.173 |
-0.251 |
| 327 |
253219_at |
AT4G34990
|
AtMYB32, myb domain protein 32, MYB32, myb domain protein 32 |
-0.172 |
-0.290 |
| 328 |
255726_at |
AT1G25530
|
[Transmembrane amino acid transporter family protein] |
-0.172 |
-0.319 |
| 329 |
249988_at |
AT5G18310
|
unknown |
-0.172 |
-0.105 |
| 330 |
265344_at |
AT2G22660
|
unknown |
-0.172 |
-0.052 |
| 331 |
266555_at |
AT2G46270
|
GBF3, G-box binding factor 3 |
-0.171 |
-0.090 |
| 332 |
258921_at |
AT3G10500
|
ANAC053, NAC domain containing protein 53, NAC053, NAC domain containing protein 53, NTL4, NAC with transmembrane motif 1-like 4 |
-0.171 |
-0.008 |
| 333 |
248092_at |
AT5G55170
|
ATSUMO3, SUM3, SMALL UBIQUITIN-LIKE MODIFIER 3, SUMO 3, SMALL UBIQUITIN-LIKE MODIFIER 3, SUMO3, small ubiquitin-like modifier 3 |
-0.170 |
-0.329 |
| 334 |
258002_at |
AT3G28930
|
AIG2, AVRRPT2-INDUCED GENE 2 |
-0.168 |
-0.211 |
| 335 |
260451_at |
AT1G72360
|
AtERF73, ERF73, ethylene response factor 73, HRE1, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 1 |
-0.168 |
-0.371 |
| 336 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
-0.168 |
-0.148 |
| 337 |
267288_at |
AT2G23680
|
[Cold acclimation protein WCOR413 family] |
-0.167 |
-0.551 |
| 338 |
251426_at |
AT3G60180
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.167 |
-0.233 |
| 339 |
261339_at |
AT1G35710
|
[Protein kinase family protein with leucine-rich repeat domain] |
-0.166 |
-0.082 |
| 340 |
259340_at |
AT3G03870
|
unknown |
-0.165 |
-0.308 |
| 341 |
260078_at |
AT1G73790
|
AtGIP2, GIP1b, GCP3-Interacting Protein 1b, GIP2, GCP3-interacting protein 2 |
-0.165 |
-0.313 |
| 342 |
246943_at |
AT5G25440
|
[Protein kinase superfamily protein] |
-0.164 |
-0.278 |
| 343 |
251137_at |
AT5G01300
|
[PEBP (phosphatidylethanolamine-binding protein) family protein] |
-0.163 |
0.006 |
| 344 |
265499_at |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
-0.162 |
0.163 |
| 345 |
267490_at |
AT2G19130
|
[S-locus lectin protein kinase family protein] |
-0.161 |
-0.338 |
| 346 |
256244_at |
AT3G12520
|
SULTR4;2, sulfate transporter 4;2 |
-0.161 |
0.382 |
| 347 |
260033_at |
AT1G68760
|
ATNUDT1, ARABIDOPSIS THALIANA NUDIX HYDROLASE HOMOLOG 1, ATNUDX1, nudix hydrolase 1, NUDX1, nudix hydrolase 1, NUDX1, NUDIX HYDROLASE 1 |
-0.161 |
-0.318 |
| 348 |
251393_at |
AT3G60640
|
ATG8G, AUTOPHAGY 8G |
-0.160 |
-0.352 |
| 349 |
258608_at |
AT3G03020
|
unknown |
-0.160 |
-0.558 |
| 350 |
266365_at |
AT2G41220
|
GLU2, glutamate synthase 2 |
-0.159 |
0.033 |
| 351 |
248061_at |
AT5G55340
|
[MBOAT (membrane bound O-acyl transferase) family protein] |
-0.158 |
-0.037 |
| 352 |
253992_at |
AT4G26060
|
[Ribosomal protein L18ae family] |
-0.158 |
-0.248 |
| 353 |
246384_at |
AT1G77370
|
[Glutaredoxin family protein] |
-0.157 |
-0.452 |
| 354 |
255360_at |
AT4G03960
|
AtPFA-DSP4, PFA-DSP4, plant and fungi atypical dual-specificity phosphatase 4 |
-0.157 |
-0.326 |
| 355 |
262237_at |
AT1G48320
|
DHNAT1, DHNA-CoA thioesterase 1 |
-0.156 |
-0.570 |
| 356 |
249806_at |
AT5G23850
|
unknown |
-0.156 |
0.262 |
| 357 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
-0.155 |
0.127 |
| 358 |
256922_at |
AT3G19010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.155 |
-0.079 |
| 359 |
253525_at |
AT4G31330
|
unknown |
-0.154 |
-0.360 |
| 360 |
261221_at |
AT1G19960
|
unknown |
-0.154 |
-0.025 |
| 361 |
255590_at |
AT4G01610
|
[Cysteine proteinases superfamily protein] |
-0.154 |
-0.000 |
| 362 |
251859_at |
AT3G54680
|
[proteophosphoglycan-related] |
-0.153 |
-0.149 |
| 363 |
253066_at |
AT4G37770
|
ACS8, 1-amino-cyclopropane-1-carboxylate synthase 8 |
-0.151 |
0.064 |
| 364 |
259747_at |
AT1G71170
|
[6-phosphogluconate dehydrogenase family protein] |
-0.150 |
-0.003 |
| 365 |
251594_at |
AT3G57630
|
[exostosin family protein] |
-0.149 |
-0.036 |
| 366 |
264938_at |
AT1G60610
|
[SBP (S-ribonuclease binding protein) family protein] |
-0.149 |
-0.184 |
| 367 |
250149_at |
AT5G14700
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.148 |
-0.189 |
| 368 |
253638_at |
AT4G30470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.148 |
0.060 |
| 369 |
255900_at |
AT1G17830
|
unknown |
-0.148 |
-0.051 |
| 370 |
250811_at |
AT5G05110
|
[Cystatin/monellin family protein] |
-0.147 |
-0.297 |
| 371 |
253987_at |
AT4G26270
|
PFK3, phosphofructokinase 3 |
-0.146 |
-0.037 |
| 372 |
265327_at |
AT2G18210
|
unknown |
-0.145 |
-0.371 |
| 373 |
263401_at |
AT2G04070
|
[MATE efflux family protein] |
-0.144 |
0.021 |
| 374 |
247529_at |
AT5G61520
|
[Major facilitator superfamily protein] |
-0.143 |
0.009 |
| 375 |
260129_at |
AT1G36380
|
unknown |
-0.143 |
-0.346 |
| 376 |
266371_at |
AT2G41410
|
[Calcium-binding EF-hand family protein] |
-0.143 |
-0.179 |
| 377 |
249811_at |
AT5G23760
|
[Copper transport protein family] |
-0.143 |
-0.534 |