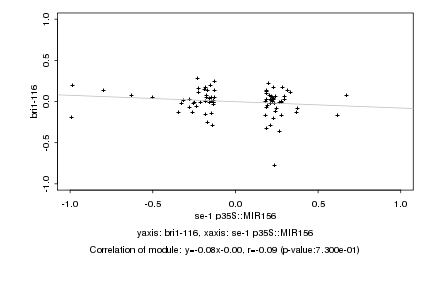
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
se-1 p35S::MIR156 |
Log2 signal ratio
bri1-116 |
| 1 |
250589_at |
AT5G07700
|
AtMYB76, myb domain protein 76, MYB76, myb domain protein 76 |
0.671 |
0.085 |
| 2 |
262719_at |
AT1G43590
|
unknown |
0.615 |
-0.168 |
| 3 |
245463_at |
AT4G17030
|
AT-EXPR, ATEXLB1, expansin-like B1, ATEXPR1, ATHEXP BETA 3.1, EXLB1, expansin-like B1, EXPR |
0.372 |
-0.075 |
| 4 |
249970_at |
AT5G19100
|
[Eukaryotic aspartyl protease family protein] |
0.366 |
-0.133 |
| 5 |
254098_at |
AT4G25100
|
ATFSD1, ARABIDOPSIS FE SUPEROXIDE DISMUTASE 1, FSD1, Fe superoxide dismutase 1 |
0.333 |
0.115 |
| 6 |
246254_at |
AT4G36450
|
ATMPK14, mitogen-activated protein kinase 14, MPK14, MPK14, mitogen-activated protein kinase 14 |
0.309 |
0.138 |
| 7 |
259423_at |
AT1G13880
|
[ELM2 domain-containing protein] |
0.295 |
0.073 |
| 8 |
265030_at |
AT1G61610
|
[S-locus lectin protein kinase family protein] |
0.293 |
0.027 |
| 9 |
265051_at |
AT1G52100
|
[Mannose-binding lectin superfamily protein] |
0.282 |
0.174 |
| 10 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
0.276 |
-0.158 |
| 11 |
248545_at |
AT5G50260
|
CEP1, cysteine endopeptidase 1 |
0.275 |
-0.005 |
| 12 |
257615_at |
AT3G26510
|
[Octicosapeptide/Phox/Bem1p family protein] |
0.273 |
0.007 |
| 13 |
253173_at |
AT4G35110
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.265 |
-0.356 |
| 14 |
248276_at |
AT5G53550
|
ATYSL3, YELLOW STRIPE LIKE 3, YSL3, YELLOW STRIPE like 3 |
0.265 |
-0.004 |
| 15 |
266555_at |
AT2G46270
|
GBF3, G-box binding factor 3 |
0.244 |
-0.075 |
| 16 |
265387_at |
AT2G20670
|
unknown |
0.242 |
0.065 |
| 17 |
263731_at |
AT1G59970
|
[Matrixin family protein] |
0.241 |
-0.113 |
| 18 |
245722_at |
AT1G73430
|
[sec34-like family protein] |
0.234 |
-0.021 |
| 19 |
256128_at |
AT1G18140
|
ATLAC1, LAC1, laccase 1 |
0.233 |
-0.768 |
| 20 |
263998_at |
AT2G22510
|
[hydroxyproline-rich glycoprotein family protein] |
0.230 |
0.042 |
| 21 |
266452_at |
AT2G43320
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.228 |
-0.201 |
| 22 |
261120_at |
AT1G75410
|
BLH3, BEL1-like homeodomain 3 |
0.226 |
0.032 |
| 23 |
265536_at |
AT2G15880
|
[Leucine-rich repeat (LRR) family protein] |
0.225 |
0.181 |
| 24 |
266313_at |
AT2G26980
|
CIPK3, CBL-interacting protein kinase 3, SnRK3.17, SNF1-RELATED PROTEIN KINASE 3.17 |
0.222 |
0.011 |
| 25 |
258890_at |
AT3G05690
|
ATHAP2B, HEME ACTIVATOR PROTEIN (YEAST) HOMOLOG 2B, HAP2B, HEME ACTIVATOR PROTEIN (YEAST) HOMOLOG 2B, NF-YA2, nuclear factor Y, subunit A2, UNE8, UNFERTILIZED EMBRYO SAC 8 |
0.220 |
0.053 |
| 26 |
255723_at |
AT3G29575
|
AFP3, ABI five binding protein 3 |
0.217 |
0.065 |
| 27 |
260205_at |
AT1G70700
|
JAZ9, JASMONATE-ZIM-DOMAIN PROTEIN 9, TIFY7 |
0.215 |
0.021 |
| 28 |
267128_at |
AT2G23620
|
ATMES1, ARABIDOPSIS THALIANA METHYL ESTERASE 1, MES1, methyl esterase 1 |
0.211 |
-0.014 |
| 29 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
0.210 |
-0.289 |
| 30 |
249645_at |
AT5G36910
|
THI2.2, thionin 2.2 |
0.207 |
0.034 |
| 31 |
254336_at |
AT4G22050
|
[Eukaryotic aspartyl protease family protein] |
0.200 |
0.079 |
| 32 |
246347_at |
AT3G56830
|
unknown |
0.195 |
0.221 |
| 33 |
260475_at |
AT1G11080
|
scpl31, serine carboxypeptidase-like 31 |
0.193 |
-0.042 |
| 34 |
256284_at |
AT3G12530
|
PSF2 |
0.187 |
0.106 |
| 35 |
250013_at |
AT5G18040
|
unknown |
0.186 |
0.122 |
| 36 |
245399_at |
AT4G17340
|
DELTA-TIP2, TIP2;2, tonoplast intrinsic protein 2;2 |
0.185 |
-0.325 |
| 37 |
247695_at |
AT5G59710
|
AtVIP2, NOT2b, Negative on TATA less2b, VIP2, VIRE2 interacting protein 2 |
0.185 |
0.033 |
| 38 |
255627_at |
AT4G00955
|
unknown |
0.183 |
-0.068 |
| 39 |
263632_at |
AT2G04795
|
unknown |
0.182 |
0.144 |
| 40 |
255825_at |
AT2G40475
|
ASG8, ALTERED SEED GERMINATION 8 |
0.181 |
-0.159 |
| 41 |
253514_at |
AT4G31805
|
POLAR, POLAR LOCALIZATION DURING ASYMMETRIC DIVISION AND REDISTRIBUTION |
0.179 |
0.007 |
| 42 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
-0.996 |
-0.183 |
| 43 |
254551_at |
AT4G19840
|
ATPP2-A1, phloem protein 2-A1, ATPP2A-1, phloem protein 2-A1, PP2-A1, phloem protein 2-A1 |
-0.991 |
0.195 |
| 44 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
-0.802 |
0.137 |
| 45 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
-0.633 |
0.079 |
| 46 |
247719_at |
AT5G59305
|
unknown |
-0.504 |
0.057 |
| 47 |
262831_at |
AT1G14730
|
[Cytochrome b561/ferric reductase transmembrane protein family] |
-0.346 |
-0.129 |
| 48 |
252096_at |
AT3G51180
|
[Zinc finger C-x8-C-x5-C-x3-H type family protein] |
-0.330 |
-0.012 |
| 49 |
247224_at |
AT5G65080
|
AGL68, AGAMOUS-like 68, MAF5, MADS AFFECTING FLOWERING 5 |
-0.320 |
0.023 |
| 50 |
256175_at |
AT1G51670
|
unknown |
-0.279 |
0.030 |
| 51 |
249619_at |
AT5G37500
|
GORK, gated outwardly-rectifying K+ channel |
-0.278 |
-0.063 |
| 52 |
255517_at |
AT4G02290
|
AtGH9B13, glycosyl hydrolase 9B13, GH9B13, glycosyl hydrolase 9B13 |
-0.263 |
-0.129 |
| 53 |
259561_at |
AT1G21250
|
AtWAK1, PRO25, WAK1, cell wall-associated kinase 1 |
-0.258 |
-0.015 |
| 54 |
254193_at |
AT4G23870
|
unknown |
-0.250 |
-0.001 |
| 55 |
262704_at |
AT1G16530
|
ASL9, ASYMMETRIC LEAVES 2-like 9, LBD3, LOB DOMAIN-CONTAINING PROTEIN 3 |
-0.239 |
-0.060 |
| 56 |
255607_at |
AT4G01130
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.230 |
0.291 |
| 57 |
262643_at |
AT1G62770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.228 |
0.117 |
| 58 |
256597_at |
AT3G28500
|
[60S acidic ribosomal protein family] |
-0.227 |
0.162 |
| 59 |
250269_at |
AT5G12970
|
[Calcium-dependent lipid-binding (CaLB domain) plant phosphoribosyltransferase family protein] |
-0.216 |
-0.001 |
| 60 |
261310_at |
AT1G05750
|
CLB19, PDE247, pigment defective 247 |
-0.188 |
0.152 |
| 61 |
258618_at |
AT3G02885
|
GASA5, GAST1 protein homolog 5 |
-0.185 |
0.006 |
| 62 |
257617_at |
AT3G26550
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.185 |
-0.147 |
| 63 |
254737_at |
AT4G13840
|
CER26, ECERIFERUM 26 |
-0.185 |
0.178 |
| 64 |
263294_at |
AT2G10870
|
unknown |
-0.180 |
0.058 |
| 65 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
-0.179 |
0.084 |
| 66 |
260515_at |
AT1G51460
|
ABCG13, ATP-binding cassette G13 |
-0.174 |
0.134 |
| 67 |
259661_at |
AT1G55265
|
unknown |
-0.171 |
-0.250 |
| 68 |
262116_at |
AT1G02816
|
unknown |
-0.159 |
0.045 |
| 69 |
267458_at |
AT2G33670
|
ATMLO5, MILDEW RESISTANCE LOCUS O 5, MLO5, MILDEW RESISTANCE LOCUS O 5 |
-0.158 |
-0.008 |
| 70 |
256713_at |
AT2G34060
|
[Peroxidase superfamily protein] |
-0.152 |
0.199 |
| 71 |
255730_at |
AT1G25460
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.149 |
0.001 |
| 72 |
260816_at |
AT1G06930
|
unknown |
-0.147 |
-0.135 |
| 73 |
247688_at |
AT5G59760
|
unknown |
-0.146 |
0.052 |
| 74 |
265032_at |
AT1G61580
|
ARP2, ARABIDOPSIS RIBOSOMAL PROTEIN 2, RPL3B, R-protein L3 B |
-0.141 |
-0.283 |
| 75 |
252285_at |
AT3G49050
|
[alpha/beta-Hydrolases superfamily protein] |
-0.136 |
-0.010 |
| 76 |
246774_at |
AT5G27530
|
[Pectin lyase-like superfamily protein] |
-0.135 |
-0.002 |
| 77 |
266177_at |
AT2G02270
|
ATPP2-B3 |
-0.134 |
0.016 |
| 78 |
251067_at |
AT5G01910
|
unknown |
-0.134 |
-0.032 |
| 79 |
252237_at |
AT3G49950
|
[GRAS family transcription factor] |
-0.132 |
0.255 |
| 80 |
245523_at |
AT4G15910
|
ATDI21, drought-induced 21, DI21, drought-induced 21 |
-0.132 |
0.141 |
| 81 |
252718_at |
AT3G43940
|
unknown |
-0.131 |
0.051 |