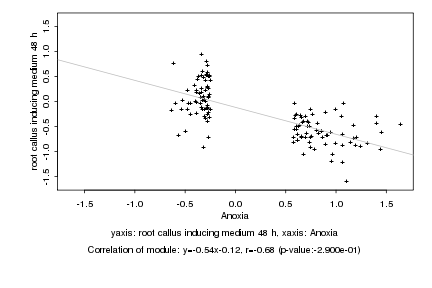
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Anoxia |
Log2 signal ratio
root callus inducing medium 48 h |
| 1 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
1.637 |
-0.450 |
| 2 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
1.444 |
-0.610 |
| 3 |
262148_at |
AT1G52560
|
[HSP20-like chaperones superfamily protein] |
1.434 |
-0.957 |
| 4 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
1.400 |
-0.284 |
| 5 |
260978_at |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
1.395 |
-0.432 |
| 6 |
253884_at |
AT4G27670
|
HSP21, heat shock protein 21 |
1.312 |
-0.836 |
| 7 |
254059_at |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
1.236 |
-0.889 |
| 8 |
262307_at |
AT1G71000
|
[Chaperone DnaJ-domain superfamily protein] |
1.199 |
-0.700 |
| 9 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
1.189 |
-0.863 |
| 10 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
1.173 |
-0.728 |
| 11 |
261838_at |
AT1G16030
|
Hsp70b, heat shock protein 70B |
1.164 |
-0.465 |
| 12 |
260522_x_at |
AT2G41730
|
unknown |
1.134 |
-0.814 |
| 13 |
259866_at |
AT1G76640
|
CML39, CALMODULIN LIKE 39 |
1.102 |
-1.594 |
| 14 |
247324_at |
AT5G64190
|
unknown |
1.072 |
-0.036 |
| 15 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
1.060 |
-0.657 |
| 16 |
266590_at |
AT2G46240
|
ATBAG6, ARABIDOPSIS THALIANA BCL-2-ASSOCIATED ATHANOGENE 6, BAG6, BCL-2-associated athanogene 6 |
1.059 |
-0.871 |
| 17 |
247208_at |
AT5G64870
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
1.056 |
-1.200 |
| 18 |
246584_at |
AT5G14730
|
unknown |
1.046 |
-0.294 |
| 19 |
248215_at |
AT5G53680
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.987 |
-0.147 |
| 20 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.985 |
-0.830 |
| 21 |
245440_at |
AT4G16680
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.961 |
-1.042 |
| 22 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
0.953 |
-1.193 |
| 23 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.939 |
-0.609 |
| 24 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
0.913 |
-0.663 |
| 25 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.902 |
-0.660 |
| 26 |
247691_at |
AT5G59720
|
HSP18.2, heat shock protein 18.2 |
0.888 |
-0.216 |
| 27 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
0.886 |
-0.851 |
| 28 |
267263_at |
AT2G23110
|
[Late embryogenesis abundant protein, group 6] |
0.863 |
-0.709 |
| 29 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
0.849 |
-0.579 |
| 30 |
259979_at |
AT1G76600
|
unknown |
0.848 |
-0.584 |
| 31 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
0.821 |
-0.624 |
| 32 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
0.811 |
-0.422 |
| 33 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.796 |
-0.563 |
| 34 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
0.779 |
-0.939 |
| 35 |
259913_at |
AT1G72660
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.760 |
-0.241 |
| 36 |
246018_at |
AT5G10695
|
unknown |
0.749 |
-0.682 |
| 37 |
263320_at |
AT2G47180
|
AtGolS1, galactinol synthase 1, GolS1, galactinol synthase 1 |
0.746 |
-0.712 |
| 38 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
0.744 |
-0.904 |
| 39 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.738 |
-0.141 |
| 40 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.729 |
-0.808 |
| 41 |
251166_at |
AT3G63350
|
AT-HSFA7B, HSFA7B, HEAT SHOCK TRANSCRIPTION FACTOR A7B |
0.728 |
-0.496 |
| 42 |
261192_at |
AT1G32870
|
ANAC013, Arabidopsis NAC domain containing protein 13, ANAC13, NAC domain protein 13, NAC13, NAC domain protein 13 |
0.720 |
-0.402 |
| 43 |
258827_at |
AT3G07150
|
unknown |
0.711 |
-0.396 |
| 44 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
0.710 |
-0.493 |
| 45 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
0.705 |
-0.622 |
| 46 |
266396_at |
AT2G38790
|
unknown |
0.693 |
-0.712 |
| 47 |
257321_at |
ATMG01130
|
ORF106F |
0.693 |
-0.288 |
| 48 |
257337_at |
ATMG00060
|
NAD5, NADH DEHYDROGENASE SUBUNIT 5, NAD5.3, NADH DEHYDROGENASE SUBUNIT 5.3, NAD5C, NADH dehydrogenase subunit 5C |
0.669 |
-0.395 |
| 49 |
253859_at |
AT4G27657
|
unknown |
0.669 |
-1.047 |
| 50 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
0.663 |
-0.413 |
| 51 |
255950_at |
AT1G22110
|
[structural constituent of ribosome] |
0.655 |
-0.708 |
| 52 |
255891_at |
AT1G17870
|
ATEGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3, EGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3 |
0.652 |
-0.301 |
| 53 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
0.650 |
-0.696 |
| 54 |
258695_at |
AT3G09640
|
APX1B, ASCORBATE PEROXIDASE 1B, APX2, ascorbate peroxidase 2 |
0.641 |
-0.275 |
| 55 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
0.633 |
-0.476 |
| 56 |
253322_at |
AT4G33980
|
unknown |
0.621 |
-0.488 |
| 57 |
257840_at |
AT3G25250
|
AGC2, AGC2-1, AGC2 kinase 1, AtOXI1, OXI1, oxidative signal-inducible1 |
0.615 |
-0.651 |
| 58 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
0.612 |
-0.771 |
| 59 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
0.608 |
-0.763 |
| 60 |
253305_at |
AT4G33666
|
unknown |
0.605 |
-0.540 |
| 61 |
258939_at |
AT3G10020
|
unknown |
0.605 |
-0.485 |
| 62 |
244950_at |
ATMG00160
|
COX2, cytochrome oxidase 2 |
0.600 |
-0.246 |
| 63 |
247543_at |
AT5G61600
|
ERF104, ethylene response factor 104 |
0.592 |
-0.263 |
| 64 |
245731_at |
AT1G73500
|
ATMKK9, MKK9, MAP kinase kinase 9 |
0.583 |
-0.025 |
| 65 |
254878_at |
AT4G11660
|
AT-HSFB2B, HSF7, HSFB2B, HEAT SHOCK TRANSCRIPTION FACTOR B2B |
0.582 |
-0.338 |
| 66 |
262656_at |
AT1G14200
|
[RING/U-box superfamily protein] |
0.578 |
-0.549 |
| 67 |
263515_at |
AT2G21640
|
unknown |
0.573 |
-0.714 |
| 68 |
259947_at |
AT1G71530
|
[Protein kinase superfamily protein] |
0.568 |
-0.802 |
| 69 |
263613_at |
AT2G25250
|
unknown |
-0.640 |
-0.172 |
| 70 |
261813_at |
AT1G08280
|
[Glycosyltransferase family 29 (sialyltransferase) family protein] |
-0.620 |
0.767 |
| 71 |
248213_at |
AT5G53660
|
AtGRF7, growth-regulating factor 7, GRF7, growth-regulating factor 7 |
-0.604 |
-0.032 |
| 72 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
-0.568 |
-0.663 |
| 73 |
261247_at |
AT1G20070
|
unknown |
-0.544 |
-0.149 |
| 74 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
-0.527 |
0.024 |
| 75 |
264470_at |
AT1G67110
|
CYP735A2, cytochrome P450, family 735, subfamily A, polypeptide 2 |
-0.502 |
-0.592 |
| 76 |
263318_at |
AT2G24762
|
AtGDU4, glutamine dumper 4, GDU4, glutamine dumper 4 |
-0.479 |
0.224 |
| 77 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.478 |
-0.154 |
| 78 |
247159_at |
AT5G65800
|
ACS5, ACC synthase 5, ATACS5, CIN5, CYTOKININ-INSENSITIVE 5, ETO2, ETHYLENE OVERPRODUCER 2 |
-0.476 |
-0.033 |
| 79 |
260367_at |
AT1G69760
|
unknown |
-0.454 |
-0.258 |
| 80 |
258003_at |
AT3G29030
|
ATEXP5, ARABIDOPSIS THALIANA EXPANSIN 5, ATEXPA5, ARABIDOPSIS THALIANA EXPANSIN A5, ATHEXP ALPHA 1.4, EXP5, EXPANSIN 5, EXPA5, expansin A5 |
-0.447 |
-0.034 |
| 81 |
263923_at |
AT2G36485
|
[ENTH/VHS family protein] |
-0.413 |
0.320 |
| 82 |
265261_at |
AT2G42990
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.404 |
0.006 |
| 83 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
-0.396 |
-0.010 |
| 84 |
251342_at |
AT3G60690
|
SAUR59, SMALL AUXIN UPREGULATED RNA 59 |
-0.390 |
-0.234 |
| 85 |
253243_at |
AT4G34560
|
unknown |
-0.388 |
0.225 |
| 86 |
257147_at |
AT3G27270
|
[TRAM, LAG1 and CLN8 (TLC) lipid-sensing domain containing protein] |
-0.388 |
0.190 |
| 87 |
257105_at |
AT3G15300
|
[VQ motif-containing protein] |
-0.383 |
0.446 |
| 88 |
246395_at |
AT1G58170
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.368 |
0.500 |
| 89 |
260301_at |
AT1G80290
|
[Nucleotide-diphospho-sugar transferases superfamily protein] |
-0.365 |
0.167 |
| 90 |
252179_at |
AT3G50760
|
GATL2, galacturonosyltransferase-like 2 |
-0.356 |
0.090 |
| 91 |
258935_at |
AT3G10120
|
unknown |
-0.355 |
0.096 |
| 92 |
264319_at |
AT1G04110
|
AtSDD1, SDD1, STOMATAL DENSITY AND DISTRIBUTION 1 |
-0.350 |
-0.023 |
| 93 |
248118_at |
AT5G55050
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.345 |
0.956 |
| 94 |
255571_at |
AT4G01220
|
MGP4, male gametophyte defective 4 |
-0.345 |
0.262 |
| 95 |
251287_at |
AT3G61820
|
[Eukaryotic aspartyl protease family protein] |
-0.339 |
0.525 |
| 96 |
250571_at |
AT5G08200
|
[peptidoglycan-binding LysM domain-containing protein] |
-0.338 |
-0.117 |
| 97 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
-0.338 |
0.190 |
| 98 |
265250_at |
AT2G01950
|
BRL2, BRI1-like 2, VH1, VASCULAR HIGHWAY 1 |
-0.337 |
0.037 |
| 99 |
260024_at |
AT1G30080
|
[Glycosyl hydrolase superfamily protein] |
-0.336 |
0.603 |
| 100 |
265283_at |
AT2G20370
|
AtMUR3, KAM1, KATAMARI 1, MUR3, MURUS 3, RSA3, SHORT ROOT IN SALT MEDIUM 3 |
-0.328 |
-0.146 |
| 101 |
250820_at |
AT5G05160
|
RUL1, REDUCED IN LATERAL GROWTH1 |
-0.326 |
0.496 |
| 102 |
254914_at |
AT4G11290
|
[Peroxidase superfamily protein] |
-0.324 |
-0.914 |
| 103 |
264770_at |
AT1G23030
|
[ARM repeat superfamily protein] |
-0.319 |
0.115 |
| 104 |
264804_at |
AT1G08590
|
[Leucine-rich receptor-like protein kinase family protein] |
-0.316 |
-0.291 |
| 105 |
254110_at |
AT4G25260
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.315 |
-0.149 |
| 106 |
259523_at |
AT1G12500
|
[Nucleotide-sugar transporter family protein] |
-0.309 |
0.042 |
| 107 |
254324_at |
AT4G22640
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.305 |
0.011 |
| 108 |
261975_at |
AT1G64640
|
AtENODL8, ENODL8, early nodulin-like protein 8 |
-0.302 |
-0.321 |
| 109 |
263241_at |
AT2G16500
|
ADC1, arginine decarboxylase 1, ARGDC, ARGDC1, SPE1 |
-0.302 |
0.231 |
| 110 |
254515_at |
AT4G20270
|
BAM3, BARELY ANY MERISTEM 3 |
-0.298 |
0.425 |
| 111 |
254284_at |
AT4G22910
|
CCS52A1, cell cycle switch protein 52 A1, FZR2, FIZZY-related 2 |
-0.296 |
-0.134 |
| 112 |
256299_at |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
-0.293 |
0.813 |
| 113 |
264527_at |
AT1G30760
|
[FAD-binding Berberine family protein] |
-0.293 |
0.530 |
| 114 |
259736_at |
AT1G64390
|
AtGH9C2, glycosyl hydrolase 9C2, GH9C2, glycosyl hydrolase 9C2 |
-0.290 |
0.546 |
| 115 |
259689_x_at |
AT1G63130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.286 |
0.584 |
| 116 |
246983_at |
AT5G67200
|
[Leucine-rich repeat protein kinase family protein] |
-0.285 |
0.738 |
| 117 |
249113_at |
AT5G43790
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.285 |
0.314 |
| 118 |
262981_at |
AT1G75590
|
SAUR52, SMALL AUXIN UPREGULATED RNA 52 |
-0.284 |
-0.241 |
| 119 |
253455_at |
AT4G32020
|
unknown |
-0.283 |
-0.137 |
| 120 |
261825_at |
AT1G11545
|
XTH8, xyloglucan endotransglucosylase/hydrolase 8 |
-0.282 |
-0.387 |
| 121 |
251254_at |
AT3G62270
|
BOR2, REQUIRES HIGH BORON 2 |
-0.281 |
-0.108 |
| 122 |
257923_at |
AT3G23160
|
unknown |
-0.280 |
-0.064 |
| 123 |
246797_at |
AT5G26790
|
unknown |
-0.279 |
0.286 |
| 124 |
259328_at |
AT3G16440
|
ATMLP-300B, myrosinase-binding protein-like protein-300B, MEE36, maternal effect embryo arrest 36, MLP-300B, myrosinase-binding protein-like protein-300B |
-0.276 |
-0.211 |
| 125 |
248006_at |
AT5G56250
|
HAP8, HAPLESS 8 |
-0.274 |
0.094 |
| 126 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
-0.273 |
0.515 |
| 127 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.272 |
-0.704 |
| 128 |
251048_at |
AT5G02410
|
ALG10, homolog of yeast ALG10 |
-0.268 |
0.263 |
| 129 |
263218_at |
AT1G30570
|
HERK2, hercules receptor kinase 2 |
-0.268 |
0.114 |
| 130 |
250015_at |
AT5G18070
|
DRT101, DNA-DAMAGE-REPAIR/TOLERATION 101 |
-0.266 |
0.158 |
| 131 |
250327_at |
AT5G12050
|
unknown |
-0.264 |
-0.314 |
| 132 |
249832_at |
AT5G23400
|
[Leucine-rich repeat (LRR) family protein] |
-0.263 |
0.510 |
| 133 |
252175_at |
AT3G50700
|
AtIDD2, indeterminate(ID)-domain 2, IDD2, indeterminate(ID)-domain 2 |
-0.258 |
0.538 |
| 134 |
248908_at |
AT5G45800
|
MEE62, maternal effect embryo arrest 62 |
-0.256 |
0.430 |
| 135 |
245412_at |
AT4G17280
|
[Auxin-responsive family protein] |
-0.254 |
-0.147 |