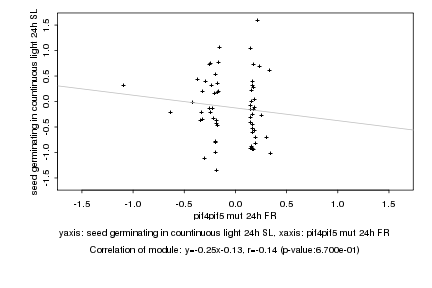
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
pif4pif5 mut 24h FR |
Log2 signal ratio
seed germinating in countinuous light 24h SL |
| 1 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
0.339 |
-1.005 |
| 2 |
265724_at |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
0.327 |
0.626 |
| 3 |
266465_at |
AT2G47750
|
GH3.9, putative indole-3-acetic acid-amido synthetase GH3.9 |
0.299 |
-0.704 |
| 4 |
263492_at |
AT2G42560
|
[late embryogenesis abundant domain-containing protein / LEA domain-containing protein] |
0.248 |
-0.271 |
| 5 |
258299_at |
AT3G23410
|
ATFAO3, ARABIDOPSIS FATTY ALCOHOL OXIDASE 3, FAO3, fatty alcohol oxidase 3 |
0.233 |
0.693 |
| 6 |
259172_at |
AT3G03630
|
CS26, cysteine synthase 26 |
0.206 |
1.609 |
| 7 |
251577_at |
AT3G58350
|
RTM3, RESTRICTED TEV MOVEMENT 3 |
0.192 |
-0.823 |
| 8 |
256024_at |
AT1G58340
|
BCD1, BUSH-AND-CHLOROTIC-DWARF 1, ZF14, ZRZ, ZRIZI |
0.190 |
-0.698 |
| 9 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.186 |
-0.549 |
| 10 |
260388_at |
AT1G74070
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
0.180 |
-0.106 |
| 11 |
260380_at |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
0.178 |
0.058 |
| 12 |
250187_at |
AT5G14370
|
[CCT motif family protein] |
0.170 |
-0.932 |
| 13 |
248723_at |
AT5G47950
|
[HXXXD-type acyl-transferase family protein] |
0.169 |
-0.148 |
| 14 |
264037_at |
AT2G03750
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.168 |
0.741 |
| 15 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
0.167 |
0.292 |
| 16 |
261596_at |
AT1G33080
|
[MATE efflux family protein] |
0.166 |
-0.243 |
| 17 |
245616_at |
AT4G14480
|
[Protein kinase superfamily protein] |
0.165 |
-0.528 |
| 18 |
245565_at |
AT4G14605
|
MDA1, MTERF DEFECTIVE IN Arabidopsis1 |
0.165 |
0.331 |
| 19 |
259082_at |
AT3G04820
|
[Pseudouridine synthase family protein] |
0.164 |
-0.903 |
| 20 |
249814_at |
AT5G23840
|
[MD-2-related lipid recognition domain-containing protein] |
0.164 |
-0.604 |
| 21 |
250915_at |
AT5G03790
|
ATHB51, HB51, homeobox 51, LMI1, LATE MERISTEM IDENTITY1 |
0.162 |
-0.440 |
| 22 |
259572_at |
AT1G20400
|
unknown |
0.162 |
-0.930 |
| 23 |
247577_at |
AT5G61290
|
[Flavin-binding monooxygenase family protein] |
0.158 |
0.402 |
| 24 |
260877_at |
AT1G21500
|
unknown |
0.153 |
0.012 |
| 25 |
254138_at |
AT4G24950
|
unknown |
0.153 |
-0.877 |
| 26 |
256020_at |
AT1G58290
|
AtHEMA1, Arabidopsis thaliana hemA 1, HEMA1 |
0.152 |
0.217 |
| 27 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
0.147 |
-0.294 |
| 28 |
251549_at |
AT3G58890
|
[RNI-like superfamily protein] |
0.146 |
-0.908 |
| 29 |
265429_at |
AT2G20710
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.143 |
-0.396 |
| 30 |
258911_at |
AT3G06470
|
[GNS1/SUR4 membrane protein family] |
0.142 |
1.058 |
| 31 |
255934_at |
AT1G12740
|
CYP87A2, cytochrome P450, family 87, subfamily A, polypeptide 2 |
0.141 |
-0.073 |
| 32 |
249439_at |
AT5G40020
|
[Pathogenesis-related thaumatin superfamily protein] |
0.141 |
-0.141 |
| 33 |
251497_at |
AT3G59060
|
PIF5, PHYTOCHROME-INTERACTING FACTOR 5, PIL6, phytochrome interacting factor 3-like 6 |
-1.099 |
0.317 |
| 34 |
254685_at |
AT4G13790
|
SAUR25, SMALL AUXIN UPREGULATED RNA 25 |
-0.639 |
-0.199 |
| 35 |
251768_at |
AT3G55940
|
[Phosphoinositide-specific phospholipase C family protein] |
-0.425 |
-0.019 |
| 36 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
-0.373 |
0.440 |
| 37 |
262198_at |
AT1G53830
|
ATPME2, pectin methylesterase 2, PME2, pectin methylesterase 2 |
-0.345 |
-0.355 |
| 38 |
261109_at |
AT1G75450
|
ATCKX5, ARABIDOPSIS THALIANA CYTOKININ OXIDASE 5, ATCKX6, CYTOKININ OXIDASE 6, CKX5, cytokinin oxidase 5 |
-0.340 |
-0.212 |
| 39 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.324 |
0.207 |
| 40 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
-0.323 |
-0.346 |
| 41 |
253155_at |
AT4G35720
|
unknown |
-0.312 |
-1.104 |
| 42 |
260841_at |
AT1G29195
|
unknown |
-0.293 |
0.405 |
| 43 |
251870_at |
AT3G54510
|
[Early-responsive to dehydration stress protein (ERD4)] |
-0.259 |
0.742 |
| 44 |
255007_at |
AT4G10020
|
AtHSD5, hydroxysteroid dehydrogenase 5, HSD5, hydroxysteroid dehydrogenase 5 |
-0.255 |
-0.129 |
| 45 |
251771_at |
AT3G56000
|
ATCSLA14, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE A14, CSLA14, cellulose synthase like A14 |
-0.249 |
-0.205 |
| 46 |
247074_at |
AT5G66590
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.246 |
0.753 |
| 47 |
261261_at |
AT1G26730
|
[EXS (ERD1/XPR1/SYG1) family protein] |
-0.241 |
0.328 |
| 48 |
264395_at |
AT1G12070
|
[Immunoglobulin E-set superfamily protein] |
-0.233 |
-0.125 |
| 49 |
249626_at |
AT5G37540
|
[Eukaryotic aspartyl protease family protein] |
-0.219 |
-0.327 |
| 50 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
-0.208 |
0.165 |
| 51 |
261046_at |
AT1G01390
|
[UDP-Glycosyltransferase superfamily protein] |
-0.203 |
-0.982 |
| 52 |
264822_at |
AT1G03457
|
AtBRN2, BRN2, Bruno-like 2 |
-0.203 |
0.537 |
| 53 |
245314_at |
AT4G16745
|
[Exostosin family protein] |
-0.201 |
-0.778 |
| 54 |
249807_at |
AT5G23870
|
[Pectinacetylesterase family protein] |
-0.198 |
-0.789 |
| 55 |
264296_at |
AT1G78720
|
[SecY protein transport family protein] |
-0.191 |
-1.335 |
| 56 |
264059_at |
AT2G31305
|
INH3, INHIBITOR-3 |
-0.187 |
-0.429 |
| 57 |
257799_at |
AT3G15890
|
[Protein kinase superfamily protein] |
-0.186 |
-0.354 |
| 58 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
-0.179 |
-0.464 |
| 59 |
247135_at |
AT5G66260
|
SAUR11, SMALL AUXIN UPREGULATED RNA 11 |
-0.179 |
0.181 |
| 60 |
267048_at |
AT2G34200
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-0.177 |
0.358 |
| 61 |
260989_at |
AT1G53450
|
unknown |
-0.168 |
0.785 |
| 62 |
250657_at |
AT5G07000
|
ATST2B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2B, ST2B, sulfotransferase 2B |
-0.166 |
0.216 |
| 63 |
249794_at |
AT5G23530
|
AtCXE18, carboxyesterase 18, CXE18, carboxyesterase 18 |
-0.161 |
1.062 |