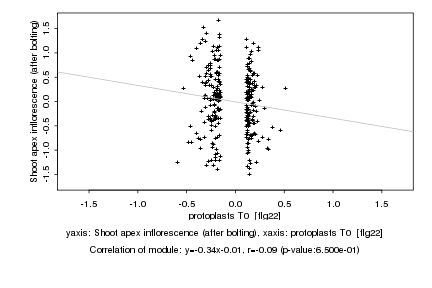
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
protoplasts T0 [flg22] |
Log2 signal ratio
Shoot apex inflorescence (after bolting) |
| 1 |
244966_at |
ATCG00600
|
PETG |
0.506 |
0.287 |
| 2 |
253582_at |
AT4G30670
|
[Putative membrane lipoprotein] |
0.457 |
-0.580 |
| 3 |
245005_at |
ATCG00330
|
RPS14, chloroplast ribosomal protein S14 |
0.371 |
-0.528 |
| 4 |
262373_at |
AT1G73120
|
unknown |
0.336 |
-0.776 |
| 5 |
259286_at |
AT3G11480
|
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
0.335 |
-0.968 |
| 6 |
260135_at |
AT1G66400
|
CML23, calmodulin like 23 |
0.327 |
-0.948 |
| 7 |
257502_at |
AT1G78110
|
unknown |
0.288 |
-0.125 |
| 8 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
0.275 |
-0.722 |
| 9 |
247902_at |
AT5G57350
|
AHA3, H(+)-ATPase 3, ATAHA3, ARABIDOPSIS THALIANA ARABIDOPSIS H(+)-ATPASE, HA3, H(+)-ATPase 3 |
0.273 |
0.300 |
| 10 |
263653_at |
AT1G04310
|
ERS2, ethylene response sensor 2 |
0.237 |
0.027 |
| 11 |
255064_at |
AT4G08950
|
EXO, EXORDIUM |
0.234 |
1.130 |
| 12 |
252023_at |
AT3G52920
|
unknown |
0.231 |
1.067 |
| 13 |
265197_at |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
0.228 |
-0.821 |
| 14 |
261333_at |
AT1G44910
|
ATPRP40A, pre-mRNA-processing protein 40A, PRP40A, pre-mRNA-processing protein 40A |
0.225 |
0.536 |
| 15 |
251985_at |
AT3G53220
|
[Thioredoxin superfamily protein] |
0.222 |
-0.409 |
| 16 |
263788_at |
AT2G24580
|
[FAD-dependent oxidoreductase family protein] |
0.213 |
-0.200 |
| 17 |
267048_at |
AT2G34200
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.208 |
0.348 |
| 18 |
260011_at |
AT1G68110
|
[ENTH/ANTH/VHS superfamily protein] |
0.206 |
-1.253 |
| 19 |
267282_at |
AT2G19390
|
unknown |
0.198 |
0.289 |
| 20 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
0.197 |
-0.665 |
| 21 |
265828_at |
AT2G14520
|
unknown |
0.196 |
-0.084 |
| 22 |
253559_at |
AT4G31140
|
[O-Glycosyl hydrolases family 17 protein] |
0.195 |
0.428 |
| 23 |
246120_at |
AT5G20360
|
Phox3, Phox3 |
0.195 |
-0.651 |
| 24 |
261813_at |
AT1G08280
|
[Glycosyltransferase family 29 (sialyltransferase) family protein] |
0.195 |
0.585 |
| 25 |
247151_at |
AT5G65640
|
bHLH093, beta HLH protein 93 |
0.194 |
0.334 |
| 26 |
253137_at |
AT4G35500
|
[Protein kinase superfamily protein] |
0.192 |
-0.448 |
| 27 |
263370_at |
AT2G20500
|
unknown |
0.189 |
0.292 |
| 28 |
264938_at |
AT1G60610
|
[SBP (S-ribonuclease binding protein) family protein] |
0.185 |
-0.014 |
| 29 |
250803_at |
AT5G04980
|
[DNAse I-like superfamily protein] |
0.182 |
-0.451 |
| 30 |
255128_at |
AT4G08310
|
unknown |
0.181 |
1.207 |
| 31 |
258184_at |
AT3G21510
|
AHP1, histidine-containing phosphotransmitter 1 |
0.179 |
-0.183 |
| 32 |
263909_at |
AT2G36490
|
AtROS1, DML1, demeter-like 1, ROS1, REPRESSOR OF SILENCING1 |
0.179 |
0.245 |
| 33 |
260416_at |
AT1G69670
|
ATCUL3B, ARABIDOPSIS THALIANA CULLIN 3B, CUL3B, cullin 3B |
0.179 |
-0.307 |
| 34 |
267428_at |
AT2G34840
|
[Coatomer epsilon subunit] |
0.178 |
0.577 |
| 35 |
264948_at |
AT1G77030
|
[hydrolases, acting on acid anhydrides, in phosphorus-containing anhydrides] |
0.178 |
-0.077 |
| 36 |
246098_at |
AT5G20400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.175 |
-0.676 |
| 37 |
246537_at |
AT5G15940
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.174 |
-0.684 |
| 38 |
264083_at |
AT2G31230
|
ATERF15, ethylene-responsive element binding factor 15, ERF15, ethylene-responsive element binding factor 15 |
0.174 |
-0.027 |
| 39 |
246995_at |
AT5G67470
|
ATFH6, ARABIDOPSIS FORMIN HOMOLOG 6, FH6, formin homolog 6 |
0.172 |
0.234 |
| 40 |
267226_at |
AT2G44010
|
unknown |
0.171 |
-0.379 |
| 41 |
261960_at |
AT1G36550
|
[similar to retrotransposon protein, putative, Ty1-copia subclass [Oryza sativa (japonica cultivar-group)] (GB:ABA98367.2)] |
0.170 |
-0.354 |
| 42 |
262362_at |
AT1G72840
|
[Disease resistance protein (TIR-NBS-LRR class)] |
0.169 |
-0.382 |
| 43 |
256513_at |
AT1G31480
|
SGR2, SHOOT GRAVITROPISM 2 |
0.168 |
0.371 |
| 44 |
256199_at |
AT1G58250
|
HPS4, hypersensitive to Pi starvation 4, SAB, SABRE |
0.166 |
0.551 |
| 45 |
264257_at |
AT1G09230
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.163 |
0.160 |
| 46 |
254185_at |
AT4G23990
|
ATCSLG3, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE-LIKE G3, CSLG3, cellulose synthase like G3 |
0.163 |
0.087 |
| 47 |
247954_at |
AT5G56870
|
BGAL4, beta-galactosidase 4 |
0.162 |
-0.687 |
| 48 |
264851_at |
AT2G17290
|
ATCDPK3, ARABIDOPSIS THALIANA CALMODULIN-DOMAIN PROTEIN KINASE 3, ATCPK6, ARABIDOPSIS THALIANA CALCIUM-DEPENDENT PROTEIN KINASE 6, CPK6, calcium dependent protein kinase 6 |
0.161 |
0.255 |
| 49 |
249811_at |
AT5G23760
|
[Copper transport protein family] |
0.160 |
1.045 |
| 50 |
256538_x_at |
AT3G32900
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT3G47260.1)] |
0.159 |
-0.749 |
| 51 |
261596_at |
AT1G33080
|
[MATE efflux family protein] |
0.158 |
-0.069 |
| 52 |
266032_x_at |
AT2G05890
|
unknown |
0.156 |
0.220 |
| 53 |
265885_at |
AT2G42330
|
[GC-rich sequence DNA-binding factor-like protein with Tuftelin interacting domain] |
0.156 |
0.839 |
| 54 |
257230_at |
AT3G16540
|
DEG11, degradation of periplasmic proteins 11, DegP11, DegP protease 11 |
0.154 |
-0.696 |
| 55 |
256394_at |
AT3G06290
|
AtSAC3B, SAC3B, yeast Sac3 homolog B |
0.151 |
0.656 |
| 56 |
255607_at |
AT4G01130
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.151 |
0.982 |
| 57 |
260605_at |
AT2G43780
|
unknown |
0.149 |
-0.219 |
| 58 |
252165_at |
AT3G50550
|
unknown |
0.148 |
0.114 |
| 59 |
245228_at |
AT3G29810
|
COBL2, COBRA-like protein 2 precursor |
0.148 |
0.630 |
| 60 |
264288_at |
AT1G62045
|
unknown |
0.148 |
0.389 |
| 61 |
264431_at |
AT1G61700
|
[RNA polymerases N / 8 kDa subunit] |
0.147 |
-1.258 |
| 62 |
246261_at |
AT1G31810
|
AFH14, Formin Homology 14 |
0.146 |
0.470 |
| 63 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.146 |
0.105 |
| 64 |
253432_at |
AT4G32450
|
MEF8S, MEF8 similar |
0.144 |
-0.451 |
| 65 |
248479_at |
AT5G50910
|
unknown |
0.143 |
-1.481 |
| 66 |
265767_at |
AT2G48110
|
MED33B, REF4, REDUCED EPIDERMAL FLUORESCENCE 4 |
0.143 |
-0.421 |
| 67 |
262741_at |
AT1G28760
|
[Uncharacterized conserved protein (DUF2215)] |
0.142 |
0.148 |
| 68 |
249527_at |
AT5G38710
|
[Methylenetetrahydrofolate reductase family protein] |
0.141 |
-0.246 |
| 69 |
258441_at |
AT3G17260
|
[hAT-like transposase family (hobo/Ac/Tam3), has a 2.6e-27 P-value blast match to GB:CAA29005 ORFa of Maize Ac (hAT-element) (Zea mays)] |
0.140 |
-1.207 |
| 70 |
252468_at |
AT3G46970
|
ATPHS2, Arabidopsis thaliana alpha-glucan phosphorylase 2, PHS2, alpha-glucan phosphorylase 2 |
0.139 |
0.783 |
| 71 |
248579_at |
AT5G49880
|
AtMAD1, MAD1, mitotic arrest deficient 1, NES1, NITRIC OXIDE-INDUCED EARLY COTYLEDON SENESCENCE 1 |
0.139 |
0.293 |
| 72 |
256613_at |
AT3G29290
|
emb2076, embryo defective 2076 |
0.138 |
0.065 |
| 73 |
253939_at |
AT4G26930
|
AtMYB97, myb domain protein 97, MYB97, myb domain protein 97 |
0.136 |
-1.359 |
| 74 |
247185_at |
AT5G65460
|
KAC2, kinesin like protein for actin based chloroplast movement 2, KCA2, kinesin CDKA ;1 associated 2 |
0.136 |
0.893 |
| 75 |
263947_at |
AT2G35820
|
[ureidoglycolate hydrolases] |
0.136 |
-0.745 |
| 76 |
258603_at |
AT3G02990
|
ATHSFA1E, heat shock transcription factor A1E, HSFA1E, heat shock transcription factor A1E |
0.136 |
0.357 |
| 77 |
264521_at |
AT1G10020
|
unknown |
0.135 |
0.255 |
| 78 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.134 |
0.694 |
| 79 |
262773_at |
AT1G13220
|
CRWN2, CROWDED NUCLEI 2, LINC2, LITTLE NUCLEI2 |
0.132 |
0.476 |
| 80 |
262787_at |
AT1G10730
|
AP1M1, adaptor protein-1 mu-adaptin 1 |
0.132 |
0.420 |
| 81 |
248099_at |
AT5G55300
|
MGO1, MGOUN 1, TOP1, TOPOISOMERASE 1, TOP1ALPHA, DNA topoisomerase I alpha |
0.132 |
0.406 |
| 82 |
261317_at |
AT1G53030
|
[Cytochrome C oxidase copper chaperone (COX17)] |
0.132 |
0.678 |
| 83 |
252021_at |
AT3G53070
|
[Putative membrane lipoprotein] |
0.132 |
-1.045 |
| 84 |
261073_at |
AT1G07300
|
[josephin protein-related] |
0.131 |
-0.847 |
| 85 |
263920_at |
AT2G36410
|
unknown |
0.131 |
0.389 |
| 86 |
267536_at |
AT2G42010
|
PLDBETA, PLDBETA1, phospholipase D beta 1 |
0.129 |
-0.476 |
| 87 |
259351_at |
AT3G05150
|
[Major facilitator superfamily protein] |
0.129 |
-0.160 |
| 88 |
245453_at |
AT4G16900
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.129 |
-0.346 |
| 89 |
261300_at |
AT1G48560
|
unknown |
0.128 |
-0.294 |
| 90 |
262269_at |
AT1G42393
|
unknown |
0.128 |
-0.761 |
| 91 |
258001_at |
AT3G28950
|
[AIG2-like (avirulence induced gene) family protein] |
0.127 |
-0.145 |
| 92 |
248278_at |
AT5G52890
|
[AT hook motif-containing protein] |
0.127 |
0.375 |
| 93 |
264550_at |
AT1G09450
|
AtHaspin, Haspin, Haspin-related gene |
0.126 |
-0.499 |
| 94 |
254254_at |
AT4G23330
|
unknown |
0.126 |
0.206 |
| 95 |
259730_at |
AT1G77660
|
[Histone H3 K4-specific methyltransferase SET7/9 family protein] |
0.126 |
-0.268 |
| 96 |
253331_at |
AT4G33490
|
[Eukaryotic aspartyl protease family protein] |
0.125 |
0.099 |
| 97 |
246326_at |
AT1G16590
|
ATREV7, REV7 |
0.125 |
0.224 |
| 98 |
254492_at |
AT4G20260
|
ATPCAP1, ARABIDOPSIS THALIANA PLASMA-MEMBRANE ASSOCIATED CATION-BINDING PROTEIN 1, MDP25, microtubule-destabilizing protein 25, PCAP1, plasma-membrane associated cation-binding protein 1 |
0.125 |
0.892 |
| 99 |
266947_at |
AT2G18830
|
unknown |
0.124 |
-0.994 |
| 100 |
254618_at |
AT4G18780
|
ATCESA8, CELLULOSE SYNTHASE 8, CESA8, CELLULOSE SYNTHASE 8, IRX1, IRREGULAR XYLEM 1, LEW2, LEAF WILTING 2 |
0.124 |
0.035 |
| 101 |
260884_at |
AT1G29240
|
unknown |
0.123 |
-0.586 |
| 102 |
251629_at |
AT3G57410
|
ATVLN3, VLN3, villin 3 |
0.123 |
0.153 |
| 103 |
261133_at |
AT1G19715
|
[Mannose-binding lectin superfamily protein] |
0.123 |
-0.227 |
| 104 |
261610_at |
AT1G49560
|
[Homeodomain-like superfamily protein] |
0.122 |
-0.214 |
| 105 |
258849_at |
AT3G03250
|
AtUGP1, UDP-GLUCOSE PYROPHOSPHORYLASE 1, UGP, UDP-glucose pyrophosphorylase, UGP1, UDP-GLUCOSE PYROPHOSPHORYLASE 1 |
0.122 |
0.320 |
| 106 |
255736_at |
AT1G25380
|
ATNDT2, ARABIDOPSIS THALIANA NAD+ TRANSPORTER 2, NDT2, NAD+ transporter 2 |
0.121 |
-0.401 |
| 107 |
250906_at |
AT5G03650
|
SBE2.2, starch branching enzyme 2.2 |
0.121 |
0.200 |
| 108 |
256519_at |
AT1G66110
|
unknown |
0.121 |
-1.323 |
| 109 |
266727_at |
AT2G03150
|
emb1579, embryo defective 1579 |
0.120 |
0.186 |
| 110 |
261698_at |
AT1G32630
|
unknown |
0.120 |
0.098 |
| 111 |
261494_at |
AT1G28420
|
HB-1, homeobox-1, RLT1, RINGLET 1 |
0.120 |
-0.462 |
| 112 |
260206_at |
AT1G70740
|
[Protein kinase superfamily protein] |
0.119 |
-0.287 |
| 113 |
254174_at |
AT4G24120
|
ATYSL1, YELLOW STRIPE LIKE 1, YSL1, YELLOW STRIPE like 1 |
0.119 |
0.555 |
| 114 |
244949_at |
ATMG00150
|
ORF116 |
0.119 |
0.025 |
| 115 |
253648_at |
AT4G29940
|
PRHA, pathogenesis related homeodomain protein A |
0.118 |
-0.932 |
| 116 |
265344_at |
AT2G22660
|
unknown |
0.118 |
0.499 |
| 117 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
0.118 |
-0.848 |
| 118 |
266990_at |
AT2G39190
|
ATATH8 |
0.117 |
-0.173 |
| 119 |
245528_at |
AT4G15530
|
PPDK, pyruvate orthophosphate dikinase |
0.117 |
-0.113 |
| 120 |
260862_at |
AT1G43850
|
SEU, seuss |
0.117 |
0.738 |
| 121 |
250420_at |
AT5G11260
|
HY5, ELONGATED HYPOCOTYL 5, TED 5, REVERSAL OF THE DET PHENOTYPE 5 |
0.116 |
-0.376 |
| 122 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
0.116 |
-0.481 |
| 123 |
257007_at |
AT3G14205
|
[Phosphoinositide phosphatase family protein] |
0.116 |
-0.380 |
| 124 |
255648_at |
AT4G00910
|
[Aluminium activated malate transporter family protein] |
0.116 |
0.152 |
| 125 |
251262_at |
AT3G62080
|
[SNF7 family protein] |
0.116 |
-1.061 |
| 126 |
262158_at |
AT1G52540
|
[Protein kinase superfamily protein] |
0.115 |
0.721 |
| 127 |
267081_at |
AT2G41210
|
PIP5K5, phosphatidylinositol- 4-phosphate 5-kinase 5 |
0.115 |
0.427 |
| 128 |
251235_at |
AT3G62860
|
[alpha/beta-Hydrolases superfamily protein] |
0.115 |
-0.670 |
| 129 |
264625_at |
AT1G09020
|
ATSNF4, SNF4, homolog of yeast sucrose nonfermenting 4 |
0.115 |
-0.351 |
| 130 |
262406_at |
AT1G34670
|
AtMYB93, myb domain protein 93, MYB93, myb domain protein 93 |
0.114 |
-0.632 |
| 131 |
259065_at |
AT3G07520
|
ATGLR1.4, GLR1.4, glutamate receptor 1.4 |
0.114 |
-0.673 |
| 132 |
256792_at |
AT3G22150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.114 |
0.771 |
| 133 |
251468_at |
AT3G59290
|
[ENTH/VHS family protein] |
0.114 |
-0.404 |
| 134 |
251875_at |
AT3G54270
|
[sucrose-6F-phosphate phosphohydrolase family protein] |
0.113 |
-0.674 |
| 135 |
266157_at |
AT2G28100
|
ATFUC1, alpha-L-fucosidase 1, FUC1, alpha-L-fucosidase 1 |
0.113 |
-0.505 |
| 136 |
246044_at |
AT5G19450
|
CDPK19, calcium-dependent protein kinase 19, CPK8 |
0.113 |
0.192 |
| 137 |
265060_at |
AT1G52150
|
ATHB-15, ATHB15, CNA, CORONA, ICU4, INCURVATA 4 |
0.113 |
0.397 |
| 138 |
266166_at |
AT2G28080
|
[UDP-Glycosyltransferase superfamily protein] |
0.112 |
1.277 |
| 139 |
254578_at |
AT4G19410
|
[Pectinacetylesterase family protein] |
0.112 |
1.124 |
| 140 |
265265_at |
AT2G42900
|
[Plant basic secretory protein (BSP) family protein] |
0.111 |
0.465 |
| 141 |
248021_at |
AT5G56500
|
Cpn60beta3, chaperonin-60beta3 |
0.111 |
-0.220 |
| 142 |
248466_at |
AT5G50720
|
ATHVA22E, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE E, HVA22E, HVA22 homologue E |
0.111 |
0.082 |
| 143 |
257780_at |
AT3G27100
|
unknown |
0.111 |
-0.147 |
| 144 |
266381_at |
AT2G14670
|
ATSUC8, sucrose-proton symporter 8, SUC8, sucrose-proton symporter 8 |
0.111 |
-0.739 |
| 145 |
253618_at |
AT4G30420
|
UMAMIT34, Usually multiple acids move in and out Transporters 34 |
0.111 |
-0.319 |
| 146 |
248110_at |
AT5G55320
|
[MBOAT (membrane bound O-acyl transferase) family protein] |
0.110 |
-0.383 |
| 147 |
245240_at |
AT1G44510
|
[copia-like retrotransposon family, has a 0. P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
0.110 |
-0.069 |
| 148 |
257657_at |
AT3G13235
|
DDI1, DNA-damage inducible 1 |
0.110 |
0.204 |
| 149 |
264202_at |
AT1G22810
|
[Integrase-type DNA-binding superfamily protein] |
-0.603 |
-1.253 |
| 150 |
257919_at |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
-0.538 |
0.276 |
| 151 |
263378_at |
AT2G40180
|
ATHPP2C5, phosphatase 2C5, PP2C5, phosphatase 2C5 |
-0.486 |
-0.832 |
| 152 |
266421_at |
AT2G38540
|
ATLTP1, ARABIDOPSIS THALIANA LIPID TRANSFER PROTEIN 1, LP1, lipid transfer protein 1, LTP1, LIPID TRANSFER PROTEIN 1 |
-0.468 |
0.937 |
| 153 |
266975_at |
AT2G39380
|
ATEXO70H2, exocyst subunit exo70 family protein H2, EXO70H2, exocyst subunit exo70 family protein H2 |
-0.461 |
-0.495 |
| 154 |
252437_at |
AT3G47380
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.460 |
-0.827 |
| 155 |
264262_at |
AT1G09200
|
H3.1, histone 3.1 |
-0.441 |
0.855 |
| 156 |
266737_at |
AT2G47140
|
AtSDR5, SDR5, short-chain dehydrogenase reductase 5 |
-0.403 |
-0.650 |
| 157 |
263797_at |
AT2G24570
|
ATWRKY17, WRKY17, WRKY DNA-binding protein 17 |
-0.400 |
1.102 |
| 158 |
252045_at |
AT3G52450
|
AtPUB22, PUB22, plant U-box 22 |
-0.382 |
-0.746 |
| 159 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.376 |
0.517 |
| 160 |
245276_at |
AT4G16780
|
ATHB-2, homeobox protein 2, ATHB2, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2, HAT4, HB-2, homeobox protein 2 |
-0.364 |
-0.965 |
| 161 |
266596_at |
AT2G46150
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.362 |
-0.776 |
| 162 |
266215_at |
AT2G06850
|
EXGT-A1, endoxyloglucan transferase A1, EXT, ENDOXYLOGLUCAN TRANSFERASE, XTH4, xyloglucan endotransglucosylase/hydrolase 4 |
-0.361 |
1.213 |
| 163 |
258037_at |
AT3G21230
|
4CL5, 4-coumarate:CoA ligase 5 |
-0.350 |
-0.190 |
| 164 |
250665_at |
AT5G06980
|
LNK4, night light-inducible and clock-regulated 4 |
-0.346 |
0.392 |
| 165 |
252711_at |
AT3G43720
|
LTPG2, glycosylphosphatidylinositol-anchored lipid protein transfer 2 |
-0.344 |
1.288 |
| 166 |
248419_at |
AT5G51550
|
EXL3, EXORDIUM like 3 |
-0.336 |
1.541 |
| 167 |
247360_at |
AT5G63450
|
CYP94B1, cytochrome P450, family 94, subfamily B, polypeptide 1 |
-0.323 |
-0.733 |
| 168 |
251097_at |
AT5G01560
|
LecRK-VI.4, L-type lectin receptor kinase VI.4, LECRKA4.3, lectin receptor kinase a4.3 |
-0.321 |
-0.416 |
| 169 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
-0.319 |
0.121 |
| 170 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
-0.317 |
0.067 |
| 171 |
258055_at |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
-0.315 |
0.389 |
| 172 |
258054_at |
AT3G16240
|
AQP1, ATTIP2;1, DELTA-TIP, delta tonoplast integral protein, DELTA-TIP1, TIP2;1 |
-0.315 |
0.345 |
| 173 |
266119_at |
AT2G02100
|
LCR69, low-molecular-weight cysteine-rich 69, PDF2.2 |
-0.314 |
0.535 |
| 174 |
259892_at |
AT1G72610
|
ATGER1, A. THALIANA GERMIN-LIKE PROTEIN 1, GER1, germin-like protein 1, GLP1, GERMIN-LIKE PROTEIN 1 |
-0.314 |
1.249 |
| 175 |
256185_at |
AT1G51700
|
ADOF1, DOF zinc finger protein 1, DOF1, DOF zinc finger protein 1 |
-0.313 |
-0.115 |
| 176 |
259793_at |
AT1G64380
|
[Integrase-type DNA-binding superfamily protein] |
-0.307 |
0.138 |
| 177 |
263761_at |
AT2G21330
|
AtFBA1, FBA1, fructose-bisphosphate aldolase 1 |
-0.305 |
1.413 |
| 178 |
254328_at |
AT4G22570
|
APT3, adenine phosphoribosyl transferase 3 |
-0.301 |
0.392 |
| 179 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
-0.301 |
0.596 |
| 180 |
245318_at |
AT4G16980
|
[arabinogalactan-protein family] |
-0.300 |
0.703 |
| 181 |
256527_at |
AT1G66100
|
[Plant thionin] |
-0.298 |
-1.311 |
| 182 |
250149_at |
AT5G14700
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.290 |
0.068 |
| 183 |
255852_at |
AT1G66970
|
GDPDL1, Glycerophosphodiester phosphodiesterase (GDPD) like 1, SVL2, SHV3-like 2 |
-0.286 |
0.748 |
| 184 |
256093_at |
AT1G20823
|
[RING/U-box superfamily protein] |
-0.285 |
-0.305 |
| 185 |
246184_at |
AT5G20950
|
[Glycosyl hydrolase family protein] |
-0.285 |
0.435 |
| 186 |
245151_at |
AT2G47550
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.285 |
-0.350 |
| 187 |
259655_at |
AT1G55210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.278 |
0.658 |
| 188 |
254431_at |
AT4G20840
|
[FAD-binding Berberine family protein] |
-0.277 |
-1.225 |
| 189 |
265722_at |
AT2G40100
|
LHCB4.3, light harvesting complex photosystem II |
-0.273 |
0.337 |
| 190 |
249290_at |
AT5G41060
|
[DHHC-type zinc finger family protein] |
-0.265 |
0.565 |
| 191 |
256796_at |
AT3G22210
|
unknown |
-0.265 |
0.794 |
| 192 |
262288_at |
AT1G70760
|
CRR23, CHLORORESPIRATORY REDUCTION 23, NdhL, NADH dehydrogenase-like complex L |
-0.263 |
-0.367 |
| 193 |
251287_at |
AT3G61820
|
[Eukaryotic aspartyl protease family protein] |
-0.263 |
0.756 |
| 194 |
260803_at |
AT1G78340
|
ATGSTU22, glutathione S-transferase TAU 22, GSTU22, glutathione S-transferase TAU 22 |
-0.262 |
-0.348 |
| 195 |
252210_at |
AT3G50410
|
OBP1, OBF binding protein 1 |
-0.262 |
0.682 |
| 196 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
-0.261 |
0.517 |
| 197 |
264839_at |
AT1G03630
|
POR C, protochlorophyllide oxidoreductase C, PORC |
-0.260 |
-0.064 |
| 198 |
245840_at |
AT1G58420
|
[Uncharacterised conserved protein UCP031279] |
-0.259 |
0.036 |
| 199 |
247693_at |
AT5G59730
|
ATEXO70H7, exocyst subunit exo70 family protein H7, EXO70H7, exocyst subunit exo70 family protein H7 |
-0.257 |
0.071 |
| 200 |
250233_at |
AT5G13460
|
IQD11, IQ-domain 11 |
-0.252 |
0.348 |
| 201 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
-0.252 |
0.529 |
| 202 |
246019_at |
AT5G10690
|
[pentatricopeptide (PPR) repeat-containing protein / CBS domain-containing protein] |
-0.250 |
-0.374 |
| 203 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
-0.250 |
-0.587 |
| 204 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
-0.250 |
-0.410 |
| 205 |
253819_at |
AT4G28350
|
LecRK-VII.2, L-type lectin receptor kinase VII.2 |
-0.248 |
-1.208 |
| 206 |
253506_at |
AT4G31980
|
unknown |
-0.246 |
-0.482 |
| 207 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
-0.242 |
-0.848 |
| 208 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
-0.241 |
1.032 |
| 209 |
266799_at |
AT2G22860
|
ATPSK2, phytosulfokine 2 precursor, PSK2, phytosulfokine 2 precursor |
-0.240 |
-0.926 |
| 210 |
245447_at |
AT4G16820
|
DALL1, DAD1-Like Lipase 1, PLA-I{beta]2, phospholipase A I beta 2 |
-0.239 |
-0.271 |
| 211 |
253286_at |
AT4G34260
|
AXY8, ALTERED XYLOGLUCAN 8, FUC95A |
-0.235 |
0.141 |
| 212 |
253799_at |
AT4G28140
|
[Integrase-type DNA-binding superfamily protein] |
-0.231 |
-1.313 |
| 213 |
250826_at |
AT5G05220
|
unknown |
-0.231 |
0.054 |
| 214 |
249770_at |
AT5G24110
|
ATWRKY30, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 30, WRKY30, WRKY DNA-binding protein 30 |
-0.229 |
-0.868 |
| 215 |
249730_at |
AT5G24430
|
[Calcium-dependent protein kinase (CDPK) family protein] |
-0.229 |
1.137 |
| 216 |
261405_at |
AT1G18740
|
unknown |
-0.228 |
0.305 |
| 217 |
266246_at |
AT2G27690
|
CYP94C1, cytochrome P450, family 94, subfamily C, polypeptide 1 |
-0.226 |
-0.377 |
| 218 |
249120_at |
AT5G43750
|
NDH18, NAD(P)H dehydrogenase 18, PnsB5, Photosynthetic NDH subcomplex B 5 |
-0.224 |
-0.194 |
| 219 |
262085_at |
AT1G56060
|
unknown |
-0.221 |
-0.217 |
| 220 |
259282_at |
AT3G11430
|
ATGPAT5, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 5, GPAT5, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 5 |
-0.221 |
-0.061 |
| 221 |
248163_at |
AT5G54510
|
DFL1, DWARF IN LIGHT 1, GH3.6, Gretchen Hagen3.6 |
-0.220 |
0.874 |
| 222 |
246792_at |
AT5G27290
|
unknown |
-0.219 |
-0.063 |
| 223 |
257835_at |
AT3G25180
|
CYP82G1, cytochrome P450, family 82, subfamily G, polypeptide 1 |
-0.217 |
0.200 |
| 224 |
266277_at |
AT2G29310
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.215 |
0.957 |
| 225 |
259439_at |
AT1G01480
|
ACS2, 1-amino-cyclopropane-1-carboxylate synthase 2, AT-ACC2 |
-0.214 |
-0.752 |
| 226 |
261247_at |
AT1G20070
|
unknown |
-0.214 |
-0.503 |
| 227 |
264435_at |
AT1G10360
|
ATGSTU18, glutathione S-transferase TAU 18, GST29, GLUTATHIONE S-TRANSFERASE 29, GSTU18, glutathione S-transferase TAU 18 |
-0.214 |
-0.369 |
| 228 |
253628_at |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
-0.212 |
-0.320 |
| 229 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
-0.211 |
-1.073 |
| 230 |
258156_at |
AT3G18050
|
unknown |
-0.210 |
0.865 |
| 231 |
248404_at |
AT5G51460
|
ATTPPA, TPPA, trehalose-6-phosphate phosphatase A |
-0.210 |
-0.285 |
| 232 |
260276_at |
AT1G80450
|
[VQ motif-containing protein] |
-0.209 |
0.470 |
| 233 |
267635_at |
AT2G42220
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.209 |
0.152 |
| 234 |
255250_at |
AT4G05100
|
AtMYB74, myb domain protein 74, MYB74, myb domain protein 74 |
-0.209 |
-0.294 |
| 235 |
254413_at |
AT4G21440
|
ATM4, A. THALIANA MYB 4, ATMYB102, MYB-like 102, MYB102, MYB102, MYB-like 102 |
-0.207 |
-1.211 |
| 236 |
267515_at |
AT2G45680
|
TCP9, TCP domain protein 9 |
-0.207 |
-0.332 |
| 237 |
263937_at |
AT2G35910
|
[RING/U-box superfamily protein] |
-0.206 |
0.044 |
| 238 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-0.205 |
-1.148 |
| 239 |
254225_at |
AT4G23670
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.203 |
1.065 |
| 240 |
257939_at |
AT3G19930
|
ATSTP4, SUGAR TRANSPORTER 4, STP4, sugar transporter 4 |
-0.203 |
0.596 |
| 241 |
262456_at |
AT1G11260
|
ATSTP1, SUGAR TRANSPORTER 1, STP1, sugar transporter 1 |
-0.201 |
0.110 |
| 242 |
254652_at |
AT4G18170
|
ATWRKY28, WRKY28, WRKY DNA-binding protein 28 |
-0.199 |
-0.976 |
| 243 |
249372_at |
AT5G40760
|
G6PD6, glucose-6-phosphate dehydrogenase 6 |
-0.196 |
0.611 |
| 244 |
254608_at |
AT4G18910
|
ATNLM2, NOD26-LIKE INTRINSIC PROTEIN 2, NIP1;2, NOD26-like intrinsic protein 1;2, NLM2, NOD26-LIKE INTRINSIC PROTEIN 2 |
-0.195 |
0.085 |
| 245 |
263287_at |
AT2G36145
|
unknown |
-0.195 |
0.044 |
| 246 |
253090_at |
AT4G36360
|
BGAL3, beta-galactosidase 3 |
-0.195 |
0.228 |
| 247 |
261500_at |
AT1G28400
|
unknown |
-0.194 |
0.598 |
| 248 |
259625_at |
AT1G42970
|
GAPB, glyceraldehyde-3-phosphate dehydrogenase B subunit |
-0.194 |
0.314 |
| 249 |
264978_at |
AT1G27120
|
[Galactosyltransferase family protein] |
-0.193 |
0.286 |
| 250 |
248377_at |
AT5G51720
|
At-NEET, NEET, NEET group protein |
-0.193 |
0.617 |
| 251 |
260754_at |
AT1G49000
|
unknown |
-0.192 |
-0.139 |
| 252 |
251305_at |
AT3G62030
|
CYP20-3, cyclophilin 20-3, ROC4, rotamase CYP 4 |
-0.190 |
0.844 |
| 253 |
266203_at |
AT2G02230
|
AtPP2-B1, phloem protein 2-B1, PP2-B1, phloem protein 2-B1 |
-0.188 |
-0.734 |
| 254 |
247776_at |
AT5G58700
|
ATPLC4, PLC4, phosphatidylinositol-speciwc phospholipase C4 |
-0.186 |
0.174 |
| 255 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
-0.185 |
-1.379 |
| 256 |
260107_at |
AT1G66430
|
[pfkB-like carbohydrate kinase family protein] |
-0.184 |
1.123 |
| 257 |
248118_at |
AT5G55050
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.184 |
-1.054 |
| 258 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
-0.183 |
-0.442 |
| 259 |
249927_at |
AT5G19220
|
ADG2, ADP GLUCOSE PYROPHOSPHORYLASE 2, APL1, ADP glucose pyrophosphorylase large subunit 1 |
-0.183 |
0.065 |
| 260 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
-0.182 |
0.128 |
| 261 |
265418_at |
AT2G20880
|
AtERF53, ERF53, ERF domain 53 |
-0.182 |
0.552 |
| 262 |
254387_at |
AT4G21850
|
ATMSRB9, methionine sulfoxide reductase B9, MSRB9, methionine sulfoxide reductase B9 |
-0.181 |
-0.168 |
| 263 |
257932_at |
AT3G17040
|
HCF107, high chlorophyll fluorescent 107 |
-0.181 |
0.584 |
| 264 |
253012_at |
AT4G37900
|
unknown |
-0.180 |
-0.347 |
| 265 |
248861_at |
AT5G46700
|
TET1, TETRASPANIN 1, TRN2, TORNADO 2 |
-0.180 |
0.012 |
| 266 |
253387_at |
AT4G33010
|
AtGLDP1, glycine decarboxylase P-protein 1, GLDP1, glycine decarboxylase P-protein 1 |
-0.179 |
0.857 |
| 267 |
248085_at |
AT5G55480
|
GDPDL4, Glycerophosphodiester phosphodiesterase (GDPD) like 4, GPDL1, glycerophosphodiesterase-like 1, SVL1, SHV3-like 1 |
-0.177 |
0.412 |
| 268 |
256275_at |
AT3G12110
|
ACT11, actin-11 |
-0.177 |
0.466 |
| 269 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
-0.176 |
-0.140 |
| 270 |
251762_at |
AT3G55800
|
SBPASE, sedoheptulose-bisphosphatase |
-0.175 |
1.687 |
| 271 |
247320_at |
AT5G64040
|
PSAN |
-0.175 |
1.067 |
| 272 |
266555_at |
AT2G46270
|
GBF3, G-box binding factor 3 |
-0.174 |
0.070 |
| 273 |
245213_at |
AT1G44575
|
CP22, NPQ4, NONPHOTOCHEMICAL QUENCHING 4, PSBS, PHOTOSYSTEM II SUBUNIT S |
-0.173 |
1.143 |
| 274 |
264837_at |
AT1G03600
|
PSB27 |
-0.173 |
1.322 |
| 275 |
250753_at |
AT5G05860
|
UGT76C2, UDP-glucosyl transferase 76C2 |
-0.172 |
-0.171 |
| 276 |
261308_at |
AT1G48480
|
RKL1, receptor-like kinase 1 |
-0.171 |
0.162 |
| 277 |
251784_at |
AT3G55330
|
PPL1, PsbP-like protein 1 |
-0.169 |
0.393 |
| 278 |
248402_at |
AT5G52100
|
CRR1, chlororespiration reduction 1 |
-0.168 |
0.135 |
| 279 |
248082_at |
AT5G55400
|
[Actin binding Calponin homology (CH) domain-containing protein] |
-0.167 |
-0.682 |
| 280 |
250937_at |
AT5G03230
|
unknown |
-0.167 |
0.596 |
| 281 |
264313_at |
AT1G70410
|
ATBCA4, BETA CARBONIC ANHYDRASE 4, BCA4, beta carbonic anhydrase 4, CA4, BETA CARBONIC ANHYDRASE 4 |
-0.167 |
-1.204 |
| 282 |
253485_at |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
-0.167 |
0.412 |
| 283 |
246532_at |
AT5G15870
|
[glycosyl hydrolase family 81 protein] |
-0.167 |
0.702 |
| 284 |
262825_at |
AT1G11790
|
ADT1, arogenate dehydratase 1, AtADT1, Arabidopsis thaliana arogenate dehydratase 1 |
-0.166 |
0.041 |
| 285 |
249037_at |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
-0.166 |
0.242 |
| 286 |
247261_at |
AT5G64460
|
[Phosphoglycerate mutase family protein] |
-0.165 |
0.884 |
| 287 |
251789_at |
AT3G55450
|
PBL1, PBS1-like 1 |
-0.164 |
1.391 |
| 288 |
258744_at |
AT3G05830
|
unknown |
-0.163 |
0.086 |
| 289 |
249230_at |
AT5G42070
|
unknown |
-0.162 |
0.355 |
| 290 |
247933_at |
AT5G56980
|
unknown |
-0.160 |
0.120 |
| 291 |
248962_at |
AT5G45680
|
ATFKBP13, FK506 BINDING PROTEIN 13, FKBP13, FK506-binding protein 13 |
-0.160 |
0.951 |
| 292 |
248814_at |
AT5G46910
|
[Transcription factor jumonji (jmj) family protein / zinc finger (C5HC2 type) family protein] |
-0.158 |
0.202 |
| 293 |
262978_at |
AT1G75780
|
TUB1, tubulin beta-1 chain |
-0.156 |
-0.330 |
| 294 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.156 |
-1.128 |
| 295 |
264773_at |
AT1G22900
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.156 |
-0.155 |