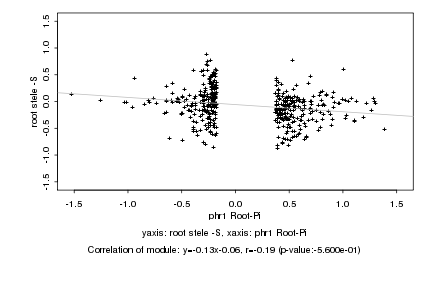
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
phr1 Root-Pi |
Log2 signal ratio
root stele -S |
| 1 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
1.386 |
-0.517 |
| 2 |
246001_at |
AT5G20790
|
unknown |
1.297 |
-0.019 |
| 3 |
263851_at |
AT2G04460
|
unknown |
1.285 |
0.007 |
| 4 |
266132_at |
AT2G45130
|
ATSPX3, ARABIDOPSIS THALIANA SPX DOMAIN GENE 3, SPX3, SPX domain gene 3 |
1.281 |
0.060 |
| 5 |
250091_at |
AT5G17340
|
[Putative membrane lipoprotein] |
1.257 |
-0.159 |
| 6 |
259399_at |
AT1G17710
|
AtPEPC1, Arabidopsis thaliana phosphoethanolamine/phosphocholine phosphatase 1, PEPC1, phosphoethanolamine/phosphocholine phosphatase 1 |
1.219 |
-0.029 |
| 7 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
1.186 |
-0.292 |
| 8 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
1.118 |
0.010 |
| 9 |
263391_at |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
1.105 |
-0.352 |
| 10 |
260522_x_at |
AT2G41730
|
unknown |
1.104 |
-0.358 |
| 11 |
261006_at |
AT1G26410
|
[FAD-binding Berberine family protein] |
1.073 |
0.068 |
| 12 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.045 |
0.005 |
| 13 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
1.026 |
-0.253 |
| 14 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
1.019 |
-0.300 |
| 15 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
1.018 |
0.022 |
| 16 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
1.005 |
0.602 |
| 17 |
258130_at |
AT3G24510
|
[Defensin-like (DEFL) family protein] |
0.996 |
0.051 |
| 18 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
0.995 |
0.046 |
| 19 |
267497_at |
AT2G30540
|
[Thioredoxin superfamily protein] |
0.959 |
-0.026 |
| 20 |
261453_at |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
0.950 |
-0.019 |
| 21 |
266983_at |
AT2G39400
|
[alpha/beta-Hydrolases superfamily protein] |
0.915 |
-0.009 |
| 22 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
0.910 |
0.183 |
| 23 |
258887_at |
AT3G05630
|
PDLZ2, PHOSPHOLIPASE D ZETA 2, PLDP2, phospholipase D P2, PLDZETA2, PHOSPHOLIPASE D ZETA 2 |
0.905 |
-0.096 |
| 24 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.899 |
0.081 |
| 25 |
261814_at |
AT1G08310
|
[alpha/beta-Hydrolases superfamily protein] |
0.894 |
-0.326 |
| 26 |
261474_at |
AT1G14540
|
PER4, peroxidase 4 |
0.887 |
-0.443 |
| 27 |
264774_at |
AT1G22890
|
unknown |
0.879 |
-0.227 |
| 28 |
261449_at |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
0.865 |
-0.169 |
| 29 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
0.845 |
0.140 |
| 30 |
254234_at |
AT4G23680
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.844 |
0.126 |
| 31 |
254772_at |
AT4G13390
|
EXT12, extensin 12 |
0.823 |
-0.056 |
| 32 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
0.815 |
-0.227 |
| 33 |
265025_at |
AT1G24575
|
unknown |
0.813 |
-0.046 |
| 34 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.808 |
0.191 |
| 35 |
250670_at |
AT5G06860
|
ATPGIP1, POLYGALACTURONASE INHIBITING PROTEIN 1, PGIP1, polygalacturonase inhibiting protein 1 |
0.808 |
-0.056 |
| 36 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.804 |
-0.331 |
| 37 |
255252_at |
AT4G04990
|
unknown |
0.793 |
0.022 |
| 38 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
0.789 |
-0.477 |
| 39 |
253044_at |
AT4G37290
|
unknown |
0.788 |
-0.188 |
| 40 |
265260_at |
AT2G43000
|
ANAC042, NAC domain containing protein 42, JUB1, JUNGBRUNNEN 1, NAC042, NAC domain containing protein 42 |
0.783 |
0.171 |
| 41 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.773 |
0.124 |
| 42 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.771 |
-0.065 |
| 43 |
258856_at |
AT3G02040
|
AtGDPD1, GDPD1, Glycerophosphodiester phosphodiesterase 1, SRG3, senescence-related gene 3 |
0.769 |
-0.070 |
| 44 |
254097_at |
AT4G25160
|
[U-box domain-containing protein kinase family protein] |
0.767 |
-0.537 |
| 45 |
247393_at |
AT5G63130
|
[Octicosapeptide/Phox/Bem1p family protein] |
0.752 |
-0.368 |
| 46 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
0.747 |
-0.165 |
| 47 |
253519_at |
AT4G31240
|
AtNRX2, NRX2, nucleoredoxin 2 |
0.717 |
0.097 |
| 48 |
255360_at |
AT4G03960
|
AtPFA-DSP4, PFA-DSP4, plant and fungi atypical dual-specificity phosphatase 4 |
0.716 |
-0.326 |
| 49 |
246370_at |
AT1G51920
|
unknown |
0.716 |
-0.171 |
| 50 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
0.713 |
-0.177 |
| 51 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.703 |
-0.046 |
| 52 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
0.700 |
0.005 |
| 53 |
264580_at |
AT1G05340
|
unknown |
0.696 |
-0.223 |
| 54 |
252730_at |
AT3G43110
|
unknown |
0.694 |
0.017 |
| 55 |
265290_at |
AT2G22590
|
[UDP-Glycosyltransferase superfamily protein] |
0.692 |
0.486 |
| 56 |
260201_at |
AT1G67600
|
[Acid phosphatase/vanadium-dependent haloperoxidase-related protein] |
0.690 |
-0.116 |
| 57 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.689 |
-0.136 |
| 58 |
265214_at |
AT1G05000
|
AtPFA-DSP1, PFA-DSP1, plant and fungi atypical dual-specificity phosphatase 1 |
0.679 |
-0.352 |
| 59 |
248752_at |
AT5G47600
|
[HSP20-like chaperones superfamily protein] |
0.675 |
0.338 |
| 60 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
0.656 |
-0.647 |
| 61 |
247729_at |
AT5G59530
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.655 |
-0.670 |
| 62 |
254524_at |
AT4G20000
|
[VQ motif-containing protein] |
0.648 |
-0.461 |
| 63 |
250515_at |
AT5G09570
|
[Cox19-like CHCH family protein] |
0.641 |
-0.122 |
| 64 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.641 |
-0.695 |
| 65 |
252334_at |
AT3G48850
|
MPT2, mitochondrial phosphate transporter 2, PHT3;2, phosphate transporter 3;2 |
0.637 |
-0.073 |
| 66 |
254014_at |
AT4G26120
|
[Ankyrin repeat family protein / BTB/POZ domain-containing protein] |
0.635 |
0.007 |
| 67 |
260239_at |
AT1G74360
|
[Leucine-rich repeat protein kinase family protein] |
0.634 |
-0.404 |
| 68 |
256009_at |
AT1G19210
|
[Integrase-type DNA-binding superfamily protein] |
0.632 |
-0.230 |
| 69 |
255958_at |
AT1G22150
|
SULTR1;3, sulfate transporter 1;3 |
0.631 |
0.055 |
| 70 |
251143_at |
AT5G01220
|
SQD2, sulfoquinovosyldiacylglycerol 2 |
0.618 |
-0.166 |
| 71 |
245875_at |
AT1G26240
|
[Proline-rich extensin-like family protein] |
0.617 |
-0.002 |
| 72 |
253859_at |
AT4G27657
|
unknown |
0.613 |
-0.269 |
| 73 |
246075_at |
AT5G20410
|
ATMGD2, ARABIDOPSIS THALIANA MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE 2, MGD2, monogalactosyldiacylglycerol synthase 2 |
0.613 |
-0.341 |
| 74 |
245082_at |
AT2G23270
|
unknown |
0.606 |
-0.278 |
| 75 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.599 |
-0.127 |
| 76 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.599 |
-0.338 |
| 77 |
245755_at |
AT1G35210
|
unknown |
0.596 |
-0.518 |
| 78 |
253657_at |
AT4G30110
|
ATHMA2, ARABIDOPSIS HEAVY METAL ATPASE 2, HMA2, heavy metal atpase 2 |
0.595 |
0.013 |
| 79 |
265008_at |
AT1G61560
|
ATMLO6, MILDEW RESISTANCE LOCUS O 6, MLO6, MILDEW RESISTANCE LOCUS O 6 |
0.587 |
-0.431 |
| 80 |
244931_at |
ATMG00630
|
ORF110B |
0.587 |
0.051 |
| 81 |
266010_at |
AT2G37430
|
ZAT11, zinc finger of Arabidopsis thaliana 11 |
0.587 |
-0.587 |
| 82 |
250499_at |
AT5G09730
|
ATBX3, ATBXL3, BETA-XYLOSIDASE 3, BX3, BXL3, beta-xylosidase 3, XYL3 |
0.585 |
0.035 |
| 83 |
249882_at |
AT5G22890
|
STOP2, sensitive to proton rhizotoxicity 2 |
0.584 |
-0.088 |
| 84 |
251026_at |
AT5G02200
|
FHL, far-red-elongated hypocotyl1-like |
0.583 |
-0.598 |
| 85 |
263515_at |
AT2G21640
|
unknown |
0.575 |
-0.300 |
| 86 |
254290_at |
AT4G23000
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
0.571 |
0.079 |
| 87 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
0.564 |
-0.644 |
| 88 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.564 |
0.310 |
| 89 |
259015_at |
AT3G07350
|
unknown |
0.562 |
0.025 |
| 90 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
0.556 |
-0.116 |
| 91 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.549 |
-0.081 |
| 92 |
258287_at |
AT3G15990
|
SULTR3;4, sulfate transporter 3;4 |
0.548 |
-0.547 |
| 93 |
247326_at |
AT5G64110
|
[Peroxidase superfamily protein] |
0.543 |
-0.018 |
| 94 |
254347_at |
AT4G22070
|
ATWRKY31, WRKY31, WRKY DNA-binding protein 31 |
0.543 |
0.111 |
| 95 |
248770_at |
AT5G47740
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.543 |
-0.357 |
| 96 |
261240_at |
AT1G32940
|
ATSBT3.5, SBT3.5 |
0.543 |
-0.015 |
| 97 |
256340_at |
AT1G72070
|
[Chaperone DnaJ-domain superfamily protein] |
0.541 |
-0.067 |
| 98 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.541 |
-0.639 |
| 99 |
249465_at |
AT5G39720
|
AIG2L, avirulence induced gene 2 like protein |
0.540 |
-0.092 |
| 100 |
251741_at |
AT3G56040
|
UGP3, UDP-glucose pyrophosphorylase 3 |
0.537 |
0.231 |
| 101 |
247617_at |
AT5G60270
|
LecRK-I.7, L-type lectin receptor kinase I.7 |
0.536 |
-0.285 |
| 102 |
253819_at |
AT4G28350
|
LecRK-VII.2, L-type lectin receptor kinase VII.2 |
0.535 |
-0.161 |
| 103 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
0.529 |
0.768 |
| 104 |
254074_at |
AT4G25490
|
ATCBF1, CBF1, C-repeat/DRE binding factor 1, DREB1B, DRE BINDING PROTEIN 1B |
0.529 |
0.063 |
| 105 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.528 |
-0.560 |
| 106 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.525 |
0.033 |
| 107 |
257615_at |
AT3G26510
|
[Octicosapeptide/Phox/Bem1p family protein] |
0.524 |
-0.383 |
| 108 |
264893_at |
AT1G23140
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.523 |
-0.668 |
| 109 |
248333_at |
AT5G52390
|
[PAR1 protein] |
0.523 |
0.003 |
| 110 |
259974_at |
AT1G76430
|
PHT1;9, phosphate transporter 1;9 |
0.521 |
-0.030 |
| 111 |
253796_at |
AT4G28460
|
unknown |
0.520 |
-0.242 |
| 112 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
0.510 |
0.033 |
| 113 |
257323_at |
ATMG01200
|
ORF294 |
0.509 |
-0.276 |
| 114 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
0.509 |
-0.712 |
| 115 |
247536_at |
AT5G61650
|
CYCP4, CYCP4;2, CYCLIN P4;2 |
0.508 |
-0.159 |
| 116 |
249750_at |
AT5G24570
|
unknown |
0.504 |
-0.467 |
| 117 |
258570_at |
AT3G04530
|
ATPPCK2, PEPCK2, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 2, PPCK2, phosphoenolpyruvate carboxylase kinase 2 |
0.503 |
0.093 |
| 118 |
265725_at |
AT2G32030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.500 |
-0.480 |
| 119 |
255753_at |
AT1G18570
|
AtMYB51, myb domain protein 51, BW51A, BW51B, HIG1, HIGH INDOLIC GLUCOSINOLATE 1, MYB51, myb domain protein 51 |
0.498 |
-0.081 |
| 120 |
261803_at |
AT1G30500
|
NF-YA7, nuclear factor Y, subunit A7 |
0.494 |
-0.048 |
| 121 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
0.494 |
-0.335 |
| 122 |
262366_at |
AT1G72890
|
[Disease resistance protein (TIR-NBS class)] |
0.492 |
-0.332 |
| 123 |
250620_at |
AT5G07190
|
ATS3, seed gene 3 |
0.491 |
-0.071 |
| 124 |
255298_at |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
0.490 |
-0.414 |
| 125 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.489 |
0.099 |
| 126 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.489 |
-0.803 |
| 127 |
259750_at |
AT1G71130
|
AtERF070, CRF8, cytokinin response factor 8, ERF070, ethylene response factor 70 |
0.487 |
-0.079 |
| 128 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
0.485 |
-0.126 |
| 129 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.484 |
-0.318 |
| 130 |
255900_at |
AT1G17830
|
unknown |
0.484 |
-0.051 |
| 131 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
0.483 |
-0.551 |
| 132 |
251652_at |
AT3G57380
|
[Glycosyltransferase family 61 protein] |
0.482 |
0.061 |
| 133 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
0.479 |
-0.709 |
| 134 |
259479_at |
AT1G19020
|
unknown |
0.479 |
-0.214 |
| 135 |
253052_at |
AT4G37310
|
CYP81H1, cytochrome P450, family 81, subfamily H, polypeptide 1 |
0.477 |
0.025 |
| 136 |
257337_at |
ATMG00060
|
NAD5, NADH DEHYDROGENASE SUBUNIT 5, NAD5.3, NADH DEHYDROGENASE SUBUNIT 5.3, NAD5C, NADH dehydrogenase subunit 5C |
0.476 |
-0.484 |
| 137 |
251507_at |
AT3G59080
|
[Eukaryotic aspartyl protease family protein] |
0.474 |
-0.321 |
| 138 |
260846_at |
AT1G17300
|
unknown |
0.473 |
-0.587 |
| 139 |
258787_at |
AT3G11840
|
PUB24, plant U-box 24 |
0.472 |
-0.297 |
| 140 |
259765_at |
AT1G64370
|
unknown |
0.471 |
0.087 |
| 141 |
251218_at |
AT3G62410
|
CP12, CP12 DOMAIN-CONTAINING PROTEIN 1, CP12-2, CP12 domain-containing protein 2 |
0.471 |
0.205 |
| 142 |
249747_at |
AT5G24600
|
unknown |
0.469 |
-0.121 |
| 143 |
247215_at |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
0.464 |
-0.706 |
| 144 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
0.462 |
-0.145 |
| 145 |
248130_at |
AT5G54790
|
unknown |
0.462 |
-0.114 |
| 146 |
259008_at |
AT3G09390
|
ATMT-1, ARABIDOPSIS THALIANA METALLOTHIONEIN-1, ATMT-K, ARABIDOPSIS THALIANA METALLOTHIONEIN-K, MT2A, metallothionein 2A |
0.460 |
-0.058 |
| 147 |
245510_at |
AT4G15740
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.460 |
0.011 |
| 148 |
247145_at |
AT5G65600
|
LecRK-IX.2, L-type lectin receptor kinase IX.2 |
0.459 |
-0.011 |
| 149 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
0.459 |
-0.274 |
| 150 |
265192_at |
AT1G05060
|
unknown |
0.458 |
-0.285 |
| 151 |
253386_at |
AT4G33030
|
SQD1, sulfoquinovosyldiacylglycerol 1 |
0.458 |
0.070 |
| 152 |
252596_at |
AT3G45330
|
LecRK-I.1, L-type lectin receptor kinase I.1 |
0.457 |
-0.208 |
| 153 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
0.457 |
-0.308 |
| 154 |
250158_at |
AT5G15190
|
unknown |
0.452 |
-0.079 |
| 155 |
250632_at |
AT5G07450
|
CYCP4;3, cyclin p4;3 |
0.452 |
-0.136 |
| 156 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
0.451 |
-0.114 |
| 157 |
259442_at |
AT1G02310
|
MAN1, endo-beta-mannanase 1 |
0.448 |
-0.213 |
| 158 |
256627_at |
AT3G19970
|
[alpha/beta-Hydrolases superfamily protein] |
0.447 |
0.032 |
| 159 |
264866_at |
AT1G24140
|
[Matrixin family protein] |
0.447 |
-0.337 |
| 160 |
261199_at |
AT1G12950
|
RSH2, root hair specific 2 |
0.445 |
-0.038 |
| 161 |
248205_at |
AT5G54300
|
unknown |
0.443 |
-0.063 |
| 162 |
248799_at |
AT5G47230
|
ATERF-5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR- 5, ATERF5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5, AtMACD1, ERF102, ERF5, ethylene responsive element binding factor 5 |
0.443 |
-0.448 |
| 163 |
264347_at |
AT1G12040
|
LRX1, leucine-rich repeat/extensin 1 |
0.440 |
-0.678 |
| 164 |
265501_at |
AT2G15490
|
UGT73B4, UDP-glycosyltransferase 73B4 |
0.438 |
0.156 |
| 165 |
246595_at |
AT5G14780
|
FDH, formate dehydrogenase |
0.435 |
-0.765 |
| 166 |
267451_at |
AT2G33710
|
[Integrase-type DNA-binding superfamily protein] |
0.435 |
-0.241 |
| 167 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
0.432 |
-0.163 |
| 168 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
0.432 |
-0.774 |
| 169 |
256697_at |
AT3G20660
|
AtOCT4, organic cation/carnitine transporter4, OCT4, organic cation/carnitine transporter4 |
0.430 |
-0.334 |
| 170 |
265387_at |
AT2G20670
|
unknown |
0.429 |
-0.302 |
| 171 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
0.429 |
0.107 |
| 172 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
0.428 |
-0.230 |
| 173 |
260611_at |
AT2G43670
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.427 |
-0.054 |
| 174 |
266017_at |
AT2G18690
|
unknown |
0.423 |
-0.239 |
| 175 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.423 |
0.325 |
| 176 |
262231_at |
AT1G68740
|
PHO1;H1 |
0.422 |
-0.149 |
| 177 |
258063_at |
AT3G14620
|
CYP72A8, cytochrome P450, family 72, subfamily A, polypeptide 8 |
0.422 |
-0.049 |
| 178 |
264940_at |
AT1G60470
|
AtGolS4, galactinol synthase 4, GolS4, galactinol synthase 4 |
0.420 |
-0.055 |
| 179 |
257832_at |
AT3G26740
|
CCL, CCR-like |
0.416 |
-0.311 |
| 180 |
265853_at |
AT2G42360
|
[RING/U-box superfamily protein] |
0.415 |
-0.281 |
| 181 |
253296_at |
AT4G33770
|
ITPK2, inositol 1,3,4-trisphosphate 5/6 kinase 2 |
0.413 |
-0.296 |
| 182 |
244901_at |
ATMG00640
|
ORF25 |
0.413 |
-0.256 |
| 183 |
248686_at |
AT5G48540
|
[receptor-like protein kinase-related family protein] |
0.412 |
-0.187 |
| 184 |
250279_at |
AT5G13200
|
[GRAM domain family protein] |
0.411 |
-0.256 |
| 185 |
266170_at |
AT2G39050
|
ArathEULS3, EULS3, Euonymus lectin S3 |
0.406 |
-0.211 |
| 186 |
263652_at |
AT1G04330
|
unknown |
0.406 |
-0.119 |
| 187 |
266658_at |
AT2G25735
|
unknown |
0.405 |
-0.382 |
| 188 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
0.404 |
0.074 |
| 189 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
0.403 |
-0.223 |
| 190 |
263987_at |
AT2G42690
|
[alpha/beta-Hydrolases superfamily protein] |
0.403 |
0.038 |
| 191 |
247047_at |
AT5G66650
|
unknown |
0.403 |
-0.155 |
| 192 |
266757_at |
AT2G46940
|
unknown |
0.401 |
-0.120 |
| 193 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
0.400 |
-0.055 |
| 194 |
253743_at |
AT4G28940
|
[Phosphorylase superfamily protein] |
0.399 |
-0.248 |
| 195 |
258151_at |
AT3G18080
|
BGLU44, B-S glucosidase 44 |
0.398 |
0.217 |
| 196 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.398 |
-0.146 |
| 197 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.398 |
-0.483 |
| 198 |
247531_at |
AT5G61550
|
[U-box domain-containing protein kinase family protein] |
0.395 |
-0.254 |
| 199 |
252437_at |
AT3G47380
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.395 |
0.154 |
| 200 |
260608_at |
AT2G43870
|
[Pectin lyase-like superfamily protein] |
0.395 |
0.236 |
| 201 |
256014_at |
AT1G19200
|
unknown |
0.395 |
0.128 |
| 202 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
0.394 |
0.371 |
| 203 |
259211_at |
AT3G09020
|
[alpha 1,4-glycosyltransferase family protein] |
0.393 |
-0.654 |
| 204 |
262745_at |
AT1G28600
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.389 |
-0.391 |
| 205 |
265796_at |
AT2G35730
|
[Heavy metal transport/detoxification superfamily protein ] |
0.388 |
-0.874 |
| 206 |
249918_at |
AT5G19240
|
[Glycoprotein membrane precursor GPI-anchored] |
0.387 |
-0.808 |
| 207 |
262324_at |
AT1G64170
|
ATCHX16, cation/H+ exchanger 16, CHX16, cation/H+ exchanger 16 |
0.386 |
-0.206 |
| 208 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
0.384 |
0.099 |
| 209 |
248298_at |
AT5G53110
|
[RING/U-box superfamily protein] |
0.382 |
-0.165 |
| 210 |
249767_at |
AT5G24090
|
ATCHIA, chitinase A, CHIA, chitinase A |
0.382 |
-0.301 |
| 211 |
253414_at |
AT4G33050
|
AtIQM1, EDA39, embryo sac development arrest 39, IQM1, IQ-motif protein 1 |
0.380 |
-0.162 |
| 212 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.379 |
0.434 |
| 213 |
259272_at |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
0.378 |
-0.133 |
| 214 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.377 |
-0.118 |
| 215 |
262505_at |
AT1G21680
|
[DPP6 N-terminal domain-like protein] |
0.377 |
0.301 |
| 216 |
257288_at |
AT3G29670
|
PMAT2, phenolic glucoside malonyltransferase 2 |
0.377 |
0.006 |
| 217 |
266109_at |
AT2G37890
|
[Mitochondrial substrate carrier family protein] |
0.376 |
0.393 |
| 218 |
249715_at |
AT5G35760
|
[Beta-galactosidase related protein] |
0.376 |
0.013 |
| 219 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
0.375 |
-0.049 |
| 220 |
252214_at |
AT3G50260
|
ATERF#011, CEJ1, cooperatively regulated by ethylene and jasmonate 1, DEAR1, DREB AND EAR MOTIF PROTEIN 1 |
0.374 |
-0.067 |
| 221 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
0.374 |
-0.145 |
| 222 |
252380_at |
AT3G47740
|
ABCA3, ATP-binding cassette A3, ATATH2, A. THALIANA ABC2 HOMOLOG 2, ATH2, ABC2 homolog 2 |
0.370 |
0.166 |
| 223 |
245560_at |
AT4G15480
|
UGT84A1 |
0.370 |
-0.348 |
| 224 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
0.370 |
-0.200 |
| 225 |
251842_at |
AT3G54580
|
[Proline-rich extensin-like family protein] |
0.369 |
-0.189 |
| 226 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
0.369 |
0.168 |
| 227 |
254550_at |
AT4G19690
|
ATIRT1, ARABIDOPSIS IRON-REGULATED TRANSPORTER 1, IRT1, iron-regulated transporter 1 |
-1.532 |
0.144 |
| 228 |
253301_at |
AT4G33720
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-1.262 |
0.030 |
| 229 |
257135_at |
AT3G12900
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-1.038 |
-0.004 |
| 230 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-1.017 |
-0.008 |
| 231 |
266222_at |
AT2G28780
|
unknown |
-0.964 |
-0.103 |
| 232 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
-0.948 |
0.440 |
| 233 |
262091_at |
AT1G56160
|
ATMYB72, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 72, MYB72, myb domain protein 72 |
-0.854 |
-0.054 |
| 234 |
250855_at |
AT5G04730
|
[Ankyrin-repeat containing protein] |
-0.814 |
0.020 |
| 235 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
-0.802 |
-0.012 |
| 236 |
252502_at |
AT3G46900
|
COPT2, copper transporter 2 |
-0.766 |
0.064 |
| 237 |
259813_at |
AT1G49860
|
ATGSTF14, glutathione S-transferase (class phi) 14, GSTF14, glutathione S-transferase (class phi) 14 |
-0.739 |
-0.034 |
| 238 |
251677_at |
AT3G56980
|
BHLH039, bHLH39, basic helix-loop-helix 39, ORG3, OBP3-RESPONSIVE GENE 3 |
-0.664 |
-0.206 |
| 239 |
251293_at |
AT3G61930
|
unknown |
-0.648 |
0.019 |
| 240 |
249535_at |
AT5G38820
|
[Transmembrane amino acid transporter family protein] |
-0.647 |
-0.194 |
| 241 |
251545_at |
AT3G58810
|
ATMTP3, ARABIDOPSIS METAL TOLERANCE PROTEIN 3, ATMTPA2, MTP3, METAL TOLERANCE PROTEIN 3, MTPA2, metal tolerance protein A2 |
-0.646 |
0.281 |
| 242 |
262373_at |
AT1G73120
|
unknown |
-0.642 |
0.031 |
| 243 |
256736_at |
AT3G29410
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.616 |
-0.689 |
| 244 |
267459_at |
AT2G33850
|
unknown |
-0.598 |
0.022 |
| 245 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
-0.590 |
-0.018 |
| 246 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
-0.588 |
0.151 |
| 247 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
-0.587 |
0.345 |
| 248 |
260783_at |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
-0.552 |
0.016 |
| 249 |
251620_at |
AT3G58060
|
[Cation efflux family protein] |
-0.544 |
0.066 |
| 250 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
-0.542 |
0.013 |
| 251 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
-0.538 |
0.056 |
| 252 |
257689_at |
AT3G12820
|
AtMYB10, myb domain protein 10, MYB10, myb domain protein 10 |
-0.521 |
-0.018 |
| 253 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
-0.511 |
-0.240 |
| 254 |
248728_at |
AT5G48000
|
CYP708 A2, CYTOCHROME P450, FAMILY 708, SUBFAMILY A, POLYPEPTIDE 2, CYP708A2, cytochrome P450, family 708, subfamily A, polypeptide 2, THAH, THALIANOL HYDROXYLASE, THAH1, THALIANOL HYDROXYLASE 1 |
-0.509 |
-0.222 |
| 255 |
260693_at |
AT1G32450
|
AtNPF7.3, NPF7.3, NRT1/ PTR family 7.3, NRT1.5, nitrate transporter 1.5 |
-0.499 |
-0.712 |
| 256 |
257153_at |
AT3G27220
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.498 |
0.095 |
| 257 |
251335_at |
AT3G61400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.496 |
-0.223 |
| 258 |
249617_at |
AT5G37450
|
[Leucine-rich repeat protein kinase family protein] |
-0.495 |
0.085 |
| 259 |
261567_at |
AT1G33055
|
unknown |
-0.482 |
-0.105 |
| 260 |
253784_at |
AT4G28610
|
AtPHR1, PHR1, phosphate starvation response 1 |
-0.480 |
0.231 |
| 261 |
264014_at |
AT2G21210
|
SAUR6, SMALL AUXIN UPREGULATED RNA 6 |
-0.477 |
0.013 |
| 262 |
248979_at |
AT5G45080
|
AtPP2-A6, phloem protein 2-A6, PP2-A6, phloem protein 2-A6 |
-0.457 |
0.046 |
| 263 |
248729_at |
AT5G48010
|
AtTHAS1, Arabidopsis thaliana thalianol synthase 1, THAS, THALIANOL SYNTHASE, THAS1, thalianol synthase 1 |
-0.447 |
-0.056 |
| 264 |
262154_at |
AT1G52700
|
[alpha/beta-Hydrolases superfamily protein] |
-0.443 |
-0.160 |
| 265 |
248727_at |
AT5G47990
|
CYP705A5, cytochrome P450, family 705, subfamily A, polypeptide 5, THAD, THALIAN-DIOL DESATURASE, THAD1, THALIAN-DIOL DESATURASE |
-0.430 |
-0.166 |
| 266 |
260883_at |
AT1G29270
|
unknown |
-0.429 |
0.115 |
| 267 |
261684_at |
AT1G47400
|
unknown |
-0.421 |
-0.290 |
| 268 |
249203_at |
AT5G42590
|
CYP71A16, cytochrome P450, family 71, subfamily A, polypeptide 16, MRO, marneral oxidase |
-0.415 |
-0.185 |
| 269 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
-0.411 |
-0.079 |
| 270 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
-0.411 |
-0.061 |
| 271 |
246142_at |
AT5G19970
|
unknown |
-0.408 |
-0.057 |
| 272 |
250828_at |
AT5G05250
|
unknown |
-0.403 |
-0.036 |
| 273 |
247603_at |
AT5G60930
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.399 |
0.596 |
| 274 |
246998_at |
AT5G67370
|
CGLD27, CONSERVED IN THE GREEN LINEAGE AND DIATOMS 27 |
-0.397 |
-0.548 |
| 275 |
254828_at |
AT4G12550
|
AIR1, Auxin-Induced in Root cultures 1 |
-0.397 |
-0.369 |
| 276 |
251761_at |
AT3G55700
|
[UDP-Glycosyltransferase superfamily protein] |
-0.392 |
-0.512 |
| 277 |
263295_at |
AT2G14210
|
AGL44, AGAMOUS-like 44, ANR1, ARABIDOPSIS NITRATE REGULATED 1 |
-0.392 |
-0.474 |
| 278 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-0.386 |
0.059 |
| 279 |
250952_at |
AT5G03570
|
ATIREG2, ARABIDOPSIS THALIANA IRON-REGULATED PROTEIN 2, FPN2, FERROPORTIN 2, IREG2, iron regulated 2 |
-0.384 |
-0.238 |
| 280 |
259228_at |
AT3G07720
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.380 |
-0.072 |
| 281 |
249144_at |
AT5G43270
|
SPL2, squamosa promoter binding protein-like 2 |
-0.379 |
0.106 |
| 282 |
253658_at |
AT4G30120
|
ATHMA3, A. THALIANA HEAVY METAL ATPASE 3, HMA3, heavy metal atpase 3 |
-0.375 |
-0.020 |
| 283 |
264305_at |
AT1G78815
|
LSH7, LIGHT SENSITIVE HYPOCOTYLS 7 |
-0.373 |
-0.146 |
| 284 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.369 |
-0.389 |
| 285 |
247577_at |
AT5G61290
|
[Flavin-binding monooxygenase family protein] |
-0.366 |
-0.551 |
| 286 |
259454_at |
AT1G44050
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.354 |
-0.632 |
| 287 |
261133_at |
AT1G19715
|
[Mannose-binding lectin superfamily protein] |
-0.337 |
-0.153 |
| 288 |
249015_at |
AT5G44730
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.333 |
0.191 |
| 289 |
249384_at |
AT5G39890
|
unknown |
-0.333 |
0.120 |
| 290 |
245586_at |
AT4G14980
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.331 |
0.132 |
| 291 |
260059_at |
AT1G78090
|
ATTPPB, Arabidopsis thaliana trehalose-6-phosphate phosphatase B, TPPB, trehalose-6-phosphate phosphatase B |
-0.330 |
-0.014 |
| 292 |
249814_at |
AT5G23840
|
[MD-2-related lipid recognition domain-containing protein] |
-0.327 |
-0.540 |
| 293 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
-0.327 |
-0.267 |
| 294 |
246557_at |
AT5G15510
|
[TPX2 (targeting protein for Xklp2) protein family] |
-0.322 |
0.579 |
| 295 |
259851_at |
AT1G72250
|
[Di-glucose binding protein with Kinesin motor domain] |
-0.317 |
0.570 |
| 296 |
264710_at |
AT1G09790
|
COBL6, COBRA-like protein 6 precursor |
-0.314 |
0.112 |
| 297 |
254013_at |
AT4G26050
|
PIRL8, plant intracellular ras group-related LRR 8 |
-0.314 |
-0.234 |
| 298 |
259151_at |
AT3G10310
|
[P-loop nucleoside triphosphate hydrolases superfamily protein with CH (Calponin Homology) domain] |
-0.314 |
0.097 |
| 299 |
265924_at |
AT2G18620
|
[Terpenoid synthases superfamily protein] |
-0.312 |
-0.166 |
| 300 |
264937_at |
AT1G61130
|
SCPL32, serine carboxypeptidase-like 32 |
-0.312 |
0.034 |
| 301 |
262905_at |
AT1G59730
|
ATH7, thioredoxin H-type 7, TH7, thioredoxin H-type 7 |
-0.309 |
-0.347 |
| 302 |
264370_at |
AT1G12100
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.306 |
-0.022 |
| 303 |
260475_at |
AT1G11080
|
scpl31, serine carboxypeptidase-like 31 |
-0.306 |
-0.080 |
| 304 |
254379_at |
AT4G21820
|
[binding] |
-0.305 |
0.596 |
| 305 |
259930_at |
AT1G34355
|
ATPS1, PARALLEL SPINDLE 1, PS1, PARALLEL SPINDLE 1 |
-0.305 |
0.500 |
| 306 |
263607_at |
AT2G16270
|
unknown |
-0.304 |
0.327 |
| 307 |
254496_at |
AT4G20070
|
AAH, allantoate amidohydrolase, ATAAH, allantoate amidohydrolase |
-0.304 |
0.023 |
| 308 |
263485_at |
AT2G29890
|
ATVLN1, VLN1, villin 1 |
-0.302 |
0.107 |
| 309 |
247024_at |
AT5G66985
|
unknown |
-0.299 |
-0.753 |
| 310 |
250152_at |
AT5G15120
|
unknown |
-0.299 |
0.110 |
| 311 |
256750_at |
AT3G27150
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.296 |
0.036 |
| 312 |
267635_at |
AT2G42220
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.295 |
0.095 |
| 313 |
260662_at |
AT1G19540
|
[NmrA-like negative transcriptional regulator family protein] |
-0.294 |
-0.142 |
| 314 |
248656_at |
AT5G48460
|
[Actin binding Calponin homology (CH) domain-containing protein] |
-0.292 |
0.131 |
| 315 |
257473_at |
AT1G33840
|
unknown |
-0.288 |
-0.083 |
| 316 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
-0.288 |
0.169 |
| 317 |
248978_at |
AT5G45070
|
AtPP2-A8, phloem protein 2-A8, PP2-A8, phloem protein 2-A8 |
-0.287 |
-0.441 |
| 318 |
256574_at |
AT3G14780
|
unknown |
-0.287 |
-0.182 |
| 319 |
259472_at |
AT1G18910
|
[zinc ion binding] |
-0.286 |
-0.167 |
| 320 |
265869_at |
AT2G01760
|
ARR14, response regulator 14, RR14, response regulator 14 |
-0.286 |
0.207 |
| 321 |
245296_at |
AT4G16370
|
oligopeptide transporter |
-0.284 |
-0.177 |
| 322 |
262628_at |
AT1G06490
|
ATGSL07, Arabidopsis thaliana glucan synthase-like 7, atgsl7, CalS7, callose synthase 7, gsl07, GSL07, glucan synthase-like 7, GSL7, glucan synthase-like 7 |
-0.281 |
0.250 |
| 323 |
248254_at |
AT5G53320
|
[Leucine-rich repeat protein kinase family protein] |
-0.280 |
-0.791 |
| 324 |
262630_at |
AT1G06520
|
ATGPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1, GPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1, sn-2-GPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1 |
-0.278 |
-0.200 |
| 325 |
256774_at |
AT3G13760
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.273 |
-0.521 |
| 326 |
259686_at |
AT1G63100
|
[GRAS family transcription factor] |
-0.273 |
0.694 |
| 327 |
247649_at |
AT5G60030
|
unknown |
-0.271 |
0.879 |
| 328 |
245227_s_at |
AT1G08410
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.270 |
0.047 |
| 329 |
250064_at |
AT5G17890
|
CHS3, CHILLING SENSITIVE 3, DAR4, DA1-related protein 4 |
-0.269 |
-0.299 |
| 330 |
253042_at |
AT4G37550
|
[Acetamidase/Formamidase family protein] |
-0.268 |
0.146 |
| 331 |
257679_at |
AT3G20470
|
ATGRP-5, ATGRP5, GRP-5, GLYCINE-RICH PROTEIN 5, GRP5, glycine-rich protein 5 |
-0.268 |
0.111 |
| 332 |
256864_at |
AT3G23890
|
ATTOPII, TOPII, topoisomerase II |
-0.267 |
0.749 |
| 333 |
258833_at |
AT3G07274
|
|
-0.266 |
0.099 |
| 334 |
267216_at |
AT2G02620
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.264 |
-0.164 |
| 335 |
255265_at |
AT4G05190
|
ATK5, kinesin 5 |
-0.263 |
0.589 |
| 336 |
261660_at |
AT1G18370
|
ATNACK1, ARABIDOPSIS NPK1-ACTIVATING KINESIN 1, HIK, HINKEL, NACK1, NPK1-ACTIVATING KINESIN 1 |
-0.261 |
0.681 |
| 337 |
246459_at |
AT5G16900
|
[Leucine-rich repeat protein kinase family protein] |
-0.260 |
-0.089 |
| 338 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
-0.259 |
0.052 |
| 339 |
260623_at |
AT1G08090
|
ACH1, ATNRT2.1, NITRATE TRANSPORTER 2.1, ATNRT2:1, nitrate transporter 2:1, LIN1, LATERAL ROOT INITIATION 1, NRT2, NITRATE TRANSPORTER 2, NRT2.1, NITRATE TRANSPORTER 2.1, NRT2:1, nitrate transporter 2:1, NRT2;1AT |
-0.259 |
-0.082 |
| 340 |
261514_at |
AT1G71870
|
[MATE efflux family protein] |
-0.257 |
-0.407 |
| 341 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
-0.257 |
-0.314 |
| 342 |
261090_at |
AT1G07560
|
[Leucine-rich repeat protein kinase family protein] |
-0.255 |
-0.393 |
| 343 |
253502_at |
AT4G31940
|
CYP82C4, cytochrome P450, family 82, subfamily C, polypeptide 4 |
-0.253 |
-0.057 |
| 344 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
-0.252 |
-0.200 |
| 345 |
264647_at |
AT1G09090
|
ATRBOHB, respiratory burst oxidase homolog B, ATRBOHB-BETA, RBOHB, respiratory burst oxidase homolog B |
-0.252 |
-0.474 |
| 346 |
248270_at |
AT5G53450
|
ORG1, OBP3-responsive gene 1 |
-0.252 |
-0.060 |
| 347 |
255550_at |
AT4G01970
|
AtSTS, stachyose synthase, RS4, raffinose synthase 4, STS, stachyose synthase |
-0.252 |
-0.007 |
| 348 |
261298_at |
AT1G48510
|
[Surfeit locus 1 cytochrome c oxidase biogenesis protein] |
-0.251 |
-0.046 |
| 349 |
246034_at |
AT5G08350
|
[GRAM domain-containing protein / ABA-responsive protein-related] |
-0.249 |
-0.215 |
| 350 |
255460_at |
AT4G02800
|
unknown |
-0.249 |
0.352 |
| 351 |
253148_at |
AT4G35620
|
CYCB2;2, Cyclin B2;2 |
-0.248 |
0.436 |
| 352 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
-0.242 |
-0.272 |
| 353 |
262114_at |
AT1G02860
|
BAH1, BENZOIC ACID HYPERSENSITIVE 1, NLA, nitrogen limitation adaptation |
-0.242 |
-0.228 |
| 354 |
254288_at |
AT4G22970
|
AESP, homolog of separase, ESP, EXTRA SPINDLE POLES, ESP, homolog of separase, RSW4, RADIALLY SWOLLEN 4 |
-0.242 |
0.417 |
| 355 |
252964_at |
AT4G38830
|
CRK26, cysteine-rich RLK (RECEPTOR-like protein kinase) 26 |
-0.241 |
-0.330 |
| 356 |
254027_at |
AT4G25835
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.241 |
-0.044 |
| 357 |
254240_at |
AT4G23496
|
SP1L5, SPIRAL1-like5 |
-0.241 |
-0.432 |
| 358 |
246962_s_at |
AT5G24800
|
ATBZIP9, ARABIDOPSIS THALIANA BASIC LEUCINE ZIPPER 9, BZIP9, basic leucine zipper 9, BZO2H2, BASIC LEUCINE ZIPPER O2 HOMOLOG 2 |
-0.241 |
-0.566 |
| 359 |
261004_at |
AT1G26450
|
[Carbohydrate-binding X8 domain superfamily protein] |
-0.240 |
-0.542 |
| 360 |
256230_at |
AT3G12340
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.240 |
0.470 |
| 361 |
261504_at |
AT1G71692
|
AGL12, AGAMOUS-like 12, XAL1, XAANTAL1 |
-0.240 |
-0.446 |
| 362 |
246054_at |
AT5G08360
|
unknown |
-0.239 |
-0.041 |
| 363 |
265414_at |
AT2G16660
|
[Major facilitator superfamily protein] |
-0.238 |
-0.077 |
| 364 |
262538_at |
AT1G17140
|
ICR1, interactor of constitutive active rops 1, RIP1, ROP INTERACTIVE PARTNER 1 |
-0.238 |
0.350 |
| 365 |
247056_at |
AT5G66750
|
ATDDM1, CHA1, CHR01, CHROMATIN REMODELING 1, CHR1, chromatin remodeling 1, DDM1, DECREASED DNA METHYLATION 1, SOM1, SOMNIFEROUS 1, SOM4 |
-0.235 |
0.772 |
| 366 |
259546_at |
AT1G35350
|
[EXS (ERD1/XPR1/SYG1) family protein] |
-0.235 |
-0.378 |
| 367 |
255236_at |
AT4G05520
|
ATEHD2, EPS15 homology domain 2, EHD2, EPS15 homology domain 2 |
-0.234 |
0.387 |
| 368 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
-0.234 |
0.082 |
| 369 |
248691_at |
AT5G48310
|
unknown |
-0.231 |
0.353 |
| 370 |
257574_at |
AT3G20710
|
[F-box family protein] |
-0.230 |
0.284 |
| 371 |
261223_at |
AT1G19950
|
HVA22H, HVA22-like protein H (ATHVA22H) |
-0.227 |
0.071 |
| 372 |
264126_at |
AT1G79280
|
AtTPR, TRANSLOCATED PROMOTER REGION, NUA, nuclear pore anchor |
-0.227 |
0.137 |
| 373 |
265667_at |
AT2G27470
|
NF-YB11, nuclear factor Y, subunit B11 |
-0.226 |
0.484 |
| 374 |
248535_at |
AT5G50120
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.226 |
-0.107 |
| 375 |
252447_at |
AT3G47040
|
[Glycosyl hydrolase family protein] |
-0.225 |
-0.018 |
| 376 |
261177_at |
AT1G04770
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.225 |
0.270 |
| 377 |
258931_at |
AT3G10010
|
DML2, demeter-like 2 |
-0.225 |
-0.161 |
| 378 |
265171_at |
AT1G23790
|
unknown |
-0.224 |
0.157 |
| 379 |
267120_at |
AT2G23530
|
[Zinc-finger domain of monoamine-oxidase A repressor R1] |
-0.223 |
0.225 |
| 380 |
260893_at |
AT1G29180
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.222 |
-0.611 |
| 381 |
247580_at |
AT5G61330
|
[rRNA processing protein-related] |
-0.222 |
0.183 |
| 382 |
256926_at |
AT3G22540
|
unknown |
-0.222 |
-0.614 |
| 383 |
246269_at |
AT4G37110
|
[Zinc-finger domain of monoamine-oxidase A repressor R1] |
-0.221 |
0.511 |
| 384 |
255955_at |
AT1G22030
|
unknown |
-0.219 |
0.074 |
| 385 |
245598_at |
AT4G14200
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.219 |
0.279 |
| 386 |
252077_at |
AT3G51720
|
unknown |
-0.219 |
0.470 |
| 387 |
253269_at |
AT4G34140
|
[D111/G-patch domain-containing protein] |
-0.219 |
0.028 |
| 388 |
247028_at |
AT5G67100
|
ICU2, INCURVATA2 |
-0.217 |
0.489 |
| 389 |
264859_at |
AT1G24280
|
G6PD3, glucose-6-phosphate dehydrogenase 3 |
-0.217 |
0.065 |
| 390 |
264939_at |
AT1G60630
|
[Leucine-rich repeat protein kinase family protein] |
-0.216 |
-0.034 |
| 391 |
264751_at |
AT1G23020
|
ATFRO3, FERRIC REDUCTION OXIDASE 3, FRO3, ferric reduction oxidase 3 |
-0.215 |
-0.216 |
| 392 |
264479_at |
AT1G77280
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.215 |
-0.018 |
| 393 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
-0.214 |
-0.083 |
| 394 |
262204_at |
AT2G01100
|
unknown |
-0.214 |
0.141 |
| 395 |
249019_at |
AT5G44780
|
MORF4, multiple organellar RNA editing factor 4 |
-0.214 |
0.325 |
| 396 |
252971_at |
AT4G38770
|
ATPRP4, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 4, PRP4, proline-rich protein 4 |
-0.213 |
0.031 |
| 397 |
265082_at |
AT1G03830
|
[guanylate-binding family protein] |
-0.213 |
0.372 |
| 398 |
258863_at |
AT3G03300
|
ATDCL2, DICER-LIKE 2, DCL2, dicer-like 2 |
-0.212 |
0.011 |
| 399 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.212 |
-0.845 |
| 400 |
258727_at |
AT3G11930
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.211 |
-0.352 |
| 401 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
-0.211 |
-0.079 |
| 402 |
261605_at |
AT1G49580
|
[Calcium-dependent protein kinase (CDPK) family protein] |
-0.209 |
-0.186 |
| 403 |
249136_at |
AT5G43180
|
unknown |
-0.209 |
-0.441 |
| 404 |
262976_at |
AT1G75520
|
SRS5, SHI-related sequence 5 |
-0.207 |
0.045 |
| 405 |
255419_at |
AT4G03230
|
[S-locus lectin protein kinase family protein] |
-0.207 |
0.053 |
| 406 |
266427_at |
AT2G07170
|
[ARM repeat superfamily protein] |
-0.206 |
0.544 |
| 407 |
257963_at |
AT3G19840
|
ATPRP40C, pre-mRNA-processing protein 40C, PRP40C, pre-mRNA-processing protein 40C |
-0.205 |
0.262 |
| 408 |
264444_at |
AT1G27360
|
SPL11, squamosa promoter-like 11 |
-0.205 |
0.098 |
| 409 |
254748_at |
AT4G13120
|
[hAT-like transposase family (hobo/Ac/Tam3), has a 2.6e-50 P-value blast match to GB:AAD24567 transposase Tag2 (hAT-element) (Arabidopsis thaliana)] |
-0.205 |
0.127 |
| 410 |
263521_at |
AT2G24960
|
unknown |
-0.204 |
0.280 |
| 411 |
257026_at |
AT3G19200
|
unknown |
-0.204 |
-0.467 |
| 412 |
245926_at |
AT5G24740
|
SHBY, SHRUBBY |
-0.204 |
-0.011 |
| 413 |
250386_at |
AT5G11510
|
AtMYB3R4, myb domain protein 3R4, MYB3R-4, myb domain protein 3r-4 |
-0.203 |
0.283 |
| 414 |
258628_at |
AT3G02890
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-0.203 |
0.296 |
| 415 |
249900_at |
AT5G22640
|
emb1211, embryo defective 1211, TIC100, TRANSLOCON AT THE INNER ENVELOPE MEMBRANE OF CHLOROPLASTS 100 |
-0.201 |
0.135 |
| 416 |
254551_at |
AT4G19840
|
ATPP2-A1, phloem protein 2-A1, ATPP2A-1, phloem protein 2-A1, PP2-A1, phloem protein 2-A1 |
-0.201 |
-0.400 |
| 417 |
259815_at |
AT1G49870
|
unknown |
-0.199 |
0.235 |
| 418 |
263194_at |
AT1G36060
|
[Integrase-type DNA-binding superfamily protein] |
-0.199 |
-0.105 |
| 419 |
262752_at |
AT1G16330
|
CYCB3;1, cyclin b3;1 |
-0.198 |
0.402 |
| 420 |
260316_at |
AT1G63810
|
unknown |
-0.198 |
0.509 |
| 421 |
256661_at |
AT3G11964
|
RRP5, Ribosomal RNA Processing 5 |
-0.197 |
0.538 |
| 422 |
249546_at |
AT5G38150
|
PMI15, plastid movement impaired 15 |
-0.196 |
0.167 |
| 423 |
245411_at |
AT4G17240
|
unknown |
-0.195 |
0.127 |
| 424 |
251154_at |
AT3G63110
|
ATIPT3, isopentenyltransferase 3, IPT3, isopentenyltransferase 3 |
-0.195 |
-0.041 |
| 425 |
262830_at |
AT1G14700
|
ATPAP3, PAP3, purple acid phosphatase 3 |
-0.194 |
-0.226 |
| 426 |
261876_at |
AT1G50590
|
[RmlC-like cupins superfamily protein] |
-0.193 |
-0.316 |
| 427 |
266805_at |
AT2G30010
|
TBL45, TRICHOME BIREFRINGENCE-LIKE 45 |
-0.193 |
-0.190 |
| 428 |
252416_at |
AT3G47460
|
ATSMC2 |
-0.193 |
0.140 |
| 429 |
252937_at |
AT4G39180
|
ATSEC14, ARABIDOPSIS THALIANA SECRETION 14, SEC14, SECRETION 14 |
-0.193 |
-0.063 |
| 430 |
251368_at |
AT3G61380
|
TRM14, TON1 Recruiting Motif 14 |
-0.193 |
-0.631 |
| 431 |
246457_at |
AT5G16750
|
TOZ, TORMOZEMBRYO DEFECTIVE |
-0.192 |
0.610 |
| 432 |
251388_at |
AT3G60840
|
MAP65-4, microtubule-associated protein 65-4 |
-0.191 |
0.159 |
| 433 |
255328_at |
AT4G04350
|
EMB2369, EMBRYO DEFECTIVE 2369 |
-0.191 |
0.084 |
| 434 |
254118_at |
AT4G24790
|
[AAA-type ATPase family protein] |
-0.191 |
0.075 |
| 435 |
250228_at |
AT5G13840
|
CCS52B, cell cycle switch protein 52 B, FZR3, FIZZY-related 3 |
-0.189 |
0.535 |
| 436 |
252478_at |
AT3G46540
|
[ENTH/VHS family protein] |
-0.187 |
0.148 |
| 437 |
257260_at |
AT3G22104
|
[Phototropic-responsive NPH3 family protein] |
-0.187 |
0.103 |
| 438 |
254466_at |
AT4G20430
|
[Subtilase family protein] |
-0.187 |
0.306 |
| 439 |
253804_at |
AT4G28230
|
unknown |
-0.186 |
0.470 |
| 440 |
250246_at |
AT5G13700
|
APAO, ATPAO1, polyamine oxidase 1, PAO1, polyamine oxidase 1 |
-0.185 |
-0.036 |
| 441 |
266187_at |
AT2G38970
|
[Zinc finger (C3HC4-type RING finger) family protein] |
-0.185 |
0.179 |
| 442 |
263415_at |
AT2G17250
|
EMB2762, EMBRYO DEFECTIVE 2762, NOC4, NucleOlar Complex associated 4 |
-0.184 |
0.364 |
| 443 |
253156_at |
AT4G35730
|
[Regulator of Vps4 activity in the MVB pathway protein] |
-0.183 |
0.303 |
| 444 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
-0.183 |
-0.579 |
| 445 |
265713_at |
AT2G03530
|
ATUPS2, ARABIDOPSIS THALIANA UREIDE PERMEASE 2, UPS2, ureide permease 2 |
-0.182 |
-0.111 |
| 446 |
264155_at |
AT1G65440
|
GTB1, global transcription factor group B1 |
-0.181 |
0.250 |
| 447 |
245461_at |
AT4G17000
|
unknown |
-0.181 |
0.277 |
| 448 |
261212_at |
AT1G12880
|
atnudt12, nudix hydrolase homolog 12, NUDT12, nudix hydrolase homolog 12 |
-0.180 |
-0.471 |
| 449 |
252736_at |
AT3G43210
|
ATNACK2, ARABIDOPSIS NPK1-ACTIVATING KINESIN 2, NACK2, NPK1-ACTIVATING KINESIN 2, TES, TETRASPORE |
-0.180 |
0.365 |
| 450 |
253213_at |
AT4G34910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.180 |
0.594 |
| 451 |
256678_at |
AT3G52380
|
CP33, chloroplast RNA-binding protein 33, PDE322, PIGMENT DEFECTIVE 322 |
-0.179 |
-0.037 |