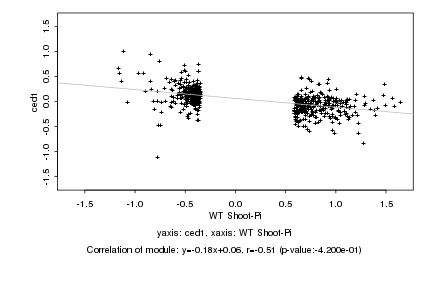
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
WT Shoot-Pi |
Log2 signal ratio
ced1 |
| 1 |
250091_at |
AT5G17340
|
[Putative membrane lipoprotein] |
1.640 |
-0.007 |
| 2 |
266132_at |
AT2G45130
|
ATSPX3, ARABIDOPSIS THALIANA SPX DOMAIN GENE 3, SPX3, SPX domain gene 3 |
1.576 |
-0.095 |
| 3 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
1.563 |
0.067 |
| 4 |
259399_at |
AT1G17710
|
AtPEPC1, Arabidopsis thaliana phosphoethanolamine/phosphocholine phosphatase 1, PEPC1, phosphoethanolamine/phosphocholine phosphatase 1 |
1.485 |
-0.068 |
| 5 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
1.478 |
0.354 |
| 6 |
250561_at |
AT5G08030
|
AtGDPD6, GDPD6, glycerophosphodiester phosphodiesterase 6 |
1.468 |
0.122 |
| 7 |
254111_at |
AT4G24890
|
ATPAP24, ARABIDOPSIS THALIANA PURPLE ACID PHOSPHATASE 24, PAP24, purple acid phosphatase 24 |
1.408 |
-0.128 |
| 8 |
266766_at |
AT2G46880
|
ATPAP14, PAP14, purple acid phosphatase 14 |
1.394 |
-0.278 |
| 9 |
259286_at |
AT3G11480
|
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
1.380 |
-0.163 |
| 10 |
246001_at |
AT5G20790
|
unknown |
1.374 |
0.027 |
| 11 |
261814_at |
AT1G08310
|
[alpha/beta-Hydrolases superfamily protein] |
1.344 |
-0.157 |
| 12 |
253087_at |
AT4G36350
|
ATPAP25, ARABIDOPSIS THALIANA PURPLE ACID PHOSPHATASE 25, PAP25, purple acid phosphatase 25 |
1.289 |
-0.027 |
| 13 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
1.282 |
0.116 |
| 14 |
258172_at |
AT3G21620
|
[ERD (early-responsive to dehydration stress) family protein] |
1.282 |
-0.065 |
| 15 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
1.272 |
-0.820 |
| 16 |
252487_at |
AT3G46660
|
UGT76E12, UDP-glucosyl transferase 76E12 |
1.223 |
-0.429 |
| 17 |
255527_at |
AT4G02360
|
unknown |
1.219 |
-0.639 |
| 18 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
1.215 |
-0.267 |
| 19 |
253832_at |
AT4G27654
|
unknown |
1.201 |
0.155 |
| 20 |
263391_at |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
1.189 |
-0.204 |
| 21 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
1.186 |
0.032 |
| 22 |
252006_at |
AT3G52820
|
ATPAP22, PURPLE ACID PHOSPHATASE 22, PAP22, purple acid phosphatase 22 |
1.184 |
-0.085 |
| 23 |
258887_at |
AT3G05630
|
PDLZ2, PHOSPHOLIPASE D ZETA 2, PLDP2, phospholipase D P2, PLDZETA2, PHOSPHOLIPASE D ZETA 2 |
1.160 |
-0.274 |
| 24 |
252730_at |
AT3G43110
|
unknown |
1.145 |
-0.085 |
| 25 |
261956_at |
AT1G64590
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
1.131 |
-0.122 |
| 26 |
249465_at |
AT5G39720
|
AIG2L, avirulence induced gene 2 like protein |
1.131 |
-0.319 |
| 27 |
267646_at |
AT2G32830
|
PHT1;5, phosphate transporter 1;5, PHT5, PHOSPHATE TRANSPORTER 5 |
1.130 |
0.055 |
| 28 |
261150_at |
AT1G19640
|
JMT, jasmonic acid carboxyl methyltransferase |
1.120 |
-0.344 |
| 29 |
247026_at |
AT5G67080
|
MAPKKK19, mitogen-activated protein kinase kinase kinase 19 |
1.116 |
-0.288 |
| 30 |
251026_at |
AT5G02200
|
FHL, far-red-elongated hypocotyl1-like |
1.113 |
0.023 |
| 31 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
1.107 |
0.056 |
| 32 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
1.106 |
-0.235 |
| 33 |
250515_at |
AT5G09570
|
[Cox19-like CHCH family protein] |
1.106 |
0.027 |
| 34 |
260140_at |
AT1G66390
|
ATMYB90, MYB90, myb domain protein 90, PAP2, PRODUCTION OF ANTHOCYANIN PIGMENT 2 |
1.101 |
-0.003 |
| 35 |
264617_at |
AT2G17660
|
[RPM1-interacting protein 4 (RIN4) family protein] |
1.101 |
-0.057 |
| 36 |
247314_at |
AT5G64000
|
ATSAL2, SAL2 |
1.100 |
-0.040 |
| 37 |
258872_at |
AT3G03260
|
HDG8, homeodomain GLABROUS 8 |
1.099 |
-0.118 |
| 38 |
258570_at |
AT3G04530
|
ATPPCK2, PEPCK2, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 2, PPCK2, phosphoenolpyruvate carboxylase kinase 2 |
1.091 |
-0.004 |
| 39 |
265122_at |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
1.091 |
-0.247 |
| 40 |
261059_at |
AT1G01250
|
[Integrase-type DNA-binding superfamily protein] |
1.086 |
-0.186 |
| 41 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
1.084 |
-0.181 |
| 42 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
1.075 |
-0.018 |
| 43 |
246075_at |
AT5G20410
|
ATMGD2, ARABIDOPSIS THALIANA MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE 2, MGD2, monogalactosyldiacylglycerol synthase 2 |
1.066 |
-0.082 |
| 44 |
260148_at |
AT1G52800
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
1.054 |
0.028 |
| 45 |
260005_at |
AT1G67920
|
unknown |
1.040 |
-0.277 |
| 46 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
1.040 |
-0.103 |
| 47 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
1.040 |
-0.031 |
| 48 |
247536_at |
AT5G61650
|
CYCP4, CYCP4;2, CYCLIN P4;2 |
1.039 |
-0.037 |
| 49 |
256340_at |
AT1G72070
|
[Chaperone DnaJ-domain superfamily protein] |
1.037 |
-0.213 |
| 50 |
259442_at |
AT1G02310
|
MAN1, endo-beta-mannanase 1 |
1.034 |
0.035 |
| 51 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
1.032 |
-0.420 |
| 52 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
1.025 |
-0.186 |
| 53 |
257645_at |
AT3G25790
|
[myb-like transcription factor family protein] |
1.018 |
-0.170 |
| 54 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
1.015 |
-0.350 |
| 55 |
254729_at |
AT4G13700
|
ATPAP23, PAP23, purple acid phosphatase 23 |
1.013 |
-0.070 |
| 56 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.004 |
0.009 |
| 57 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
1.003 |
0.259 |
| 58 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
1.001 |
0.075 |
| 59 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
0.999 |
-0.004 |
| 60 |
264886_at |
AT1G61120
|
GES, GERANYLLINALOOL SYNTHASE, TPS04, terpene synthase 04, TPS4, TERPENE SYNTHASE 4 |
0.995 |
-0.324 |
| 61 |
253859_at |
AT4G27657
|
unknown |
0.990 |
0.030 |
| 62 |
256913_at |
AT3G23870
|
unknown |
0.989 |
-0.075 |
| 63 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
0.988 |
-0.310 |
| 64 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
0.983 |
-0.630 |
| 65 |
245882_at |
AT5G09470
|
DIC3, dicarboxylate carrier 3 |
0.974 |
0.078 |
| 66 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
0.971 |
0.005 |
| 67 |
256603_at |
AT3G28270
|
unknown |
0.970 |
-0.070 |
| 68 |
265214_at |
AT1G05000
|
AtPFA-DSP1, PFA-DSP1, plant and fungi atypical dual-specificity phosphatase 1 |
0.966 |
-0.092 |
| 69 |
267075_at |
AT2G41070
|
ATBZIP12, DPBF4, EEL, ENHANCED EM LEVEL |
0.964 |
-0.247 |
| 70 |
257271_at |
AT3G28007
|
AtSWEET4, SWEET4 |
0.964 |
-0.577 |
| 71 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.964 |
-0.130 |
| 72 |
263320_at |
AT2G47180
|
AtGolS1, galactinol synthase 1, GolS1, galactinol synthase 1 |
0.959 |
-0.065 |
| 73 |
253519_at |
AT4G31240
|
AtNRX2, NRX2, nucleoredoxin 2 |
0.959 |
-0.286 |
| 74 |
255900_at |
AT1G17830
|
unknown |
0.958 |
-0.413 |
| 75 |
254097_at |
AT4G25160
|
[U-box domain-containing protein kinase family protein] |
0.955 |
-0.112 |
| 76 |
251315_at |
AT3G61410
|
unknown |
0.952 |
-0.143 |
| 77 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
0.947 |
0.082 |
| 78 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.939 |
-0.339 |
| 79 |
256014_at |
AT1G19200
|
unknown |
0.936 |
-0.281 |
| 80 |
258287_at |
AT3G15990
|
SULTR3;4, sulfate transporter 3;4 |
0.933 |
0.232 |
| 81 |
250098_at |
AT5G17350
|
unknown |
0.933 |
0.030 |
| 82 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.930 |
0.072 |
| 83 |
251012_at |
AT5G02580
|
unknown |
0.928 |
0.041 |
| 84 |
259974_at |
AT1G76430
|
PHT1;9, phosphate transporter 1;9 |
0.928 |
-0.038 |
| 85 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
0.925 |
0.441 |
| 86 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
0.925 |
-0.090 |
| 87 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
0.924 |
-0.028 |
| 88 |
264893_at |
AT1G23140
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.922 |
0.028 |
| 89 |
258932_at |
AT3G10150
|
ATPAP16, PAP16, purple acid phosphatase 16 |
0.919 |
0.092 |
| 90 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
0.919 |
-0.027 |
| 91 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
0.918 |
-0.076 |
| 92 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.916 |
-0.232 |
| 93 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
0.916 |
0.102 |
| 94 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
0.910 |
0.379 |
| 95 |
264261_at |
AT1G09240
|
ATNAS3, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 3, NAS3, nicotianamine synthase 3 |
0.908 |
0.039 |
| 96 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
0.899 |
-0.021 |
| 97 |
258322_at |
AT3G22740
|
HMT3, homocysteine S-methyltransferase 3 |
0.895 |
-0.227 |
| 98 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.892 |
-0.062 |
| 99 |
245465_at |
AT4G16590
|
ATCSLA01, cellulose synthase-like A01, ATCSLA1, CELLULOSE SYNTHASE-LIKE A1, CSLA01, CSLA01, cellulose synthase-like A01 |
0.892 |
-0.014 |
| 100 |
254683_at |
AT4G13800
|
unknown |
0.889 |
0.036 |
| 101 |
247360_at |
AT5G63450
|
CYP94B1, cytochrome P450, family 94, subfamily B, polypeptide 1 |
0.889 |
-0.138 |
| 102 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
0.886 |
0.093 |
| 103 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.885 |
-0.287 |
| 104 |
266564_at |
AT2G24000
|
scpl22, serine carboxypeptidase-like 22 |
0.883 |
-0.043 |
| 105 |
250558_at |
AT5G07990
|
CYP75B1, CYTOCHROME P450 75B1, D501, TT7, TRANSPARENT TESTA 7 |
0.875 |
-0.132 |
| 106 |
260201_at |
AT1G67600
|
[Acid phosphatase/vanadium-dependent haloperoxidase-related protein] |
0.872 |
-0.173 |
| 107 |
256924_at |
AT3G29590
|
AT5MAT |
0.870 |
-0.084 |
| 108 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
0.867 |
-0.152 |
| 109 |
253830_at |
AT4G27652
|
unknown |
0.858 |
0.154 |
| 110 |
247393_at |
AT5G63130
|
[Octicosapeptide/Phox/Bem1p family protein] |
0.857 |
-0.160 |
| 111 |
263031_at |
AT1G24070
|
ATCSLA10, cellulose synthase-like A10, CSLA10, CELLULOSE SYNTHASE LIKE A10, CSLA10, cellulose synthase-like A10 |
0.855 |
-0.175 |
| 112 |
251531_at |
AT3G58550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.854 |
-0.224 |
| 113 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
0.852 |
0.180 |
| 114 |
263998_at |
AT2G22510
|
[hydroxyproline-rich glycoprotein family protein] |
0.846 |
-0.365 |
| 115 |
256442_at |
AT3G10930
|
unknown |
0.845 |
-0.095 |
| 116 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
0.840 |
-0.002 |
| 117 |
263594_at |
AT2G01880
|
ATPAP7, PURPLE ACID PHOSPHATASE 7, PAP7, purple acid phosphatase 7 |
0.839 |
-0.165 |
| 118 |
261922_at |
AT1G65890
|
AAE12, acyl activating enzyme 12 |
0.839 |
0.191 |
| 119 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
0.837 |
0.349 |
| 120 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
0.833 |
-0.168 |
| 121 |
250292_at |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
0.832 |
0.083 |
| 122 |
256100_at |
AT1G13750
|
[Purple acid phosphatases superfamily protein] |
0.828 |
0.030 |
| 123 |
253259_at |
AT4G34410
|
RRTF1, redox responsive transcription factor 1 |
0.826 |
0.360 |
| 124 |
255958_at |
AT1G22150
|
SULTR1;3, sulfate transporter 1;3 |
0.826 |
0.065 |
| 125 |
258604_at |
AT3G02980
|
MCC1, MEIOTIC CONTROL OF CROSSOVERS1 |
0.825 |
0.003 |
| 126 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
0.823 |
-0.415 |
| 127 |
249151_at |
AT5G43360
|
ATPT4, PHT1;3, phosphate transporter 1;3, PHT3, PHOSPHATE TRANSPORTER 3 |
0.821 |
-0.002 |
| 128 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
0.815 |
-0.008 |
| 129 |
262658_at |
AT1G14220
|
[Ribonuclease T2 family protein] |
0.815 |
-0.068 |
| 130 |
263851_at |
AT2G04460
|
unknown |
0.811 |
0.126 |
| 131 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
0.803 |
-0.319 |
| 132 |
255360_at |
AT4G03960
|
AtPFA-DSP4, PFA-DSP4, plant and fungi atypical dual-specificity phosphatase 4 |
0.803 |
-0.054 |
| 133 |
247175_at |
AT5G65280
|
GCL1, GCR2-like 1 |
0.799 |
-0.171 |
| 134 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
0.797 |
-0.301 |
| 135 |
264497_at |
AT1G30840
|
ATPUP4, purine permease 4, PUP4, purine permease 4 |
0.796 |
-0.210 |
| 136 |
246342_at |
AT3G56700
|
AtFAR6, FAR6, fatty acid reductase 6 |
0.796 |
0.011 |
| 137 |
264907_at |
AT2G17280
|
[Phosphoglycerate mutase family protein] |
0.795 |
-0.078 |
| 138 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
0.794 |
-0.201 |
| 139 |
252334_at |
AT3G48850
|
MPT2, mitochondrial phosphate transporter 2, PHT3;2, phosphate transporter 3;2 |
0.793 |
-0.075 |
| 140 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.791 |
-0.439 |
| 141 |
261803_at |
AT1G30500
|
NF-YA7, nuclear factor Y, subunit A7 |
0.790 |
-0.424 |
| 142 |
256159_at |
AT1G30135
|
JAZ8, jasmonate-zim-domain protein 8, TIFY5A |
0.788 |
0.091 |
| 143 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
0.785 |
0.091 |
| 144 |
254996_at |
AT4G10390
|
[Protein kinase superfamily protein] |
0.776 |
0.056 |
| 145 |
249377_at |
AT5G40690
|
unknown |
0.776 |
-0.275 |
| 146 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.774 |
0.020 |
| 147 |
259953_at |
AT1G74810
|
BOR5, REQUIRES HIGH BORON 5 |
0.773 |
-0.260 |
| 148 |
251420_at |
AT3G60490
|
[Integrase-type DNA-binding superfamily protein] |
0.772 |
0.116 |
| 149 |
265723_at |
AT2G32140
|
[transmembrane receptors] |
0.772 |
0.045 |
| 150 |
259418_at |
AT1G02390
|
ATGPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2, GPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2 |
0.771 |
-0.095 |
| 151 |
249747_at |
AT5G24600
|
unknown |
0.769 |
-0.119 |
| 152 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
0.767 |
-0.189 |
| 153 |
265334_at |
AT2G18370
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.766 |
-0.211 |
| 154 |
255763_at |
AT1G16730
|
unknown |
0.763 |
0.207 |
| 155 |
250344_at |
AT5G11930
|
[Thioredoxin superfamily protein] |
0.761 |
0.062 |
| 156 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
0.759 |
0.028 |
| 157 |
256750_at |
AT3G27150
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.756 |
-0.025 |
| 158 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.755 |
-0.247 |
| 159 |
251143_at |
AT5G01220
|
SQD2, sulfoquinovosyldiacylglycerol 2 |
0.755 |
-0.147 |
| 160 |
260399_at |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
0.753 |
0.410 |
| 161 |
254290_at |
AT4G23000
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
0.751 |
-0.024 |
| 162 |
257592_at |
AT3G24982
|
ATRLP40, receptor like protein 40, RLP40, receptor like protein 40 |
0.750 |
0.032 |
| 163 |
265276_at |
AT2G28400
|
unknown |
0.749 |
-0.261 |
| 164 |
252266_at |
AT3G49630
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.744 |
-0.056 |
| 165 |
258856_at |
AT3G02040
|
AtGDPD1, GDPD1, Glycerophosphodiester phosphodiesterase 1, SRG3, senescence-related gene 3 |
0.743 |
-0.046 |
| 166 |
257288_at |
AT3G29670
|
PMAT2, phenolic glucoside malonyltransferase 2 |
0.739 |
-0.027 |
| 167 |
262231_at |
AT1G68740
|
PHO1;H1 |
0.739 |
0.192 |
| 168 |
256779_at |
AT3G13784
|
AtcwINV5, cell wall invertase 5, CWINV5, cell wall invertase 5 |
0.738 |
-0.103 |
| 169 |
250723_at |
AT5G06300
|
LOG7, LONELY GUY 7 |
0.737 |
-0.088 |
| 170 |
256009_at |
AT1G19210
|
[Integrase-type DNA-binding superfamily protein] |
0.737 |
0.456 |
| 171 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
0.736 |
-0.592 |
| 172 |
265197_at |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
0.731 |
-0.297 |
| 173 |
250213_at |
AT5G13820
|
ATBP-1, ATBP1, ATTBP1, HPPBF-1, H-PROTEIN PROMOTE, TBP1, telomeric DNA binding protein 1 |
0.728 |
-0.390 |
| 174 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
0.726 |
0.019 |
| 175 |
263544_at |
AT2G21590
|
APL4 |
0.725 |
0.089 |
| 176 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
0.725 |
-0.044 |
| 177 |
249134_at |
AT5G43150
|
unknown |
0.724 |
-0.104 |
| 178 |
262211_at |
AT1G74930
|
ORA47 |
0.722 |
0.464 |
| 179 |
252212_at |
AT3G50310
|
MAPKKK20, mitogen-activated protein kinase kinase kinase 20, MKKK20, MAPKK kinase 20 |
0.720 |
0.116 |
| 180 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
0.718 |
-0.156 |
| 181 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
0.716 |
-0.239 |
| 182 |
262616_at |
AT1G06620
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.714 |
-0.094 |
| 183 |
252179_at |
AT3G50760
|
GATL2, galacturonosyltransferase-like 2 |
0.712 |
0.047 |
| 184 |
260549_at |
AT2G43535
|
[Scorpion toxin-like knottin superfamily protein] |
0.711 |
0.055 |
| 185 |
256697_at |
AT3G20660
|
AtOCT4, organic cation/carnitine transporter4, OCT4, organic cation/carnitine transporter4 |
0.710 |
-0.544 |
| 186 |
260553_at |
AT2G41800
|
unknown |
0.706 |
-0.037 |
| 187 |
261037_at |
AT1G17420
|
ATLOX3, Arabidopsis thaliana lipoxygenase 3, LOX3, lipoxygenase 3 |
0.704 |
0.273 |
| 188 |
253024_at |
AT4G38080
|
[hydroxyproline-rich glycoprotein family protein] |
0.701 |
-0.187 |
| 189 |
254098_at |
AT4G25100
|
ATFSD1, ARABIDOPSIS FE SUPEROXIDE DISMUTASE 1, FSD1, Fe superoxide dismutase 1 |
0.699 |
0.069 |
| 190 |
264887_at |
AT1G23120
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.699 |
-0.029 |
| 191 |
254185_at |
AT4G23990
|
ATCSLG3, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE-LIKE G3, CSLG3, cellulose synthase like G3 |
0.696 |
0.070 |
| 192 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
0.696 |
-0.262 |
| 193 |
256654_at |
AT3G18880
|
[Nucleic acid-binding, OB-fold-like protein] |
0.696 |
0.003 |
| 194 |
245445_at |
AT4G16750
|
[Integrase-type DNA-binding superfamily protein] |
0.695 |
-0.122 |
| 195 |
264729_at |
AT1G22990
|
HIPP22, heavy metal associated isoprenylated plant protein 22 |
0.695 |
-0.355 |
| 196 |
264525_at |
AT1G10060
|
ATBCAT-1, branched-chain amino acid transaminase 1, BCAT-1, branched-chain amino acid transaminase 1, BCAT1, branched-chain amino acid transferase 1 |
0.695 |
-0.494 |
| 197 |
266765_at |
AT2G46860
|
AtPPa3, pyrophosphorylase 3, PPa3, pyrophosphorylase 3 |
0.694 |
-0.038 |
| 198 |
260230_at |
AT1G74500
|
ATBS1, activation-tagged BRI1(brassinosteroid-insensitive 1)-suppressor 1, bHLH135, basic helix-loop-helix protein 135, BS1, activation-tagged BRI1(brassinosteroid-insensitive 1)-suppressor 1, PRE3, PACLOBUTRAZOL RESISTANCE 3, TMO7, TARGET OF MONOPTEROS 7 |
0.694 |
-0.181 |
| 199 |
257644_at |
AT3G25780
|
AOC3, allene oxide cyclase 3 |
0.694 |
0.155 |
| 200 |
266669_at |
AT2G29750
|
UGT71C1, UDP-glucosyl transferase 71C1 |
0.693 |
-0.135 |
| 201 |
267496_at |
AT2G30550
|
DALL3, DAD1-Like Lipase 3 |
0.688 |
-0.123 |
| 202 |
254153_at |
AT4G24450
|
ATGWD2, GWD3, PWD, phosphoglucan, water dikinase |
0.686 |
-0.139 |
| 203 |
245799_at |
AT1G45616
|
AtRLP6, receptor like protein 6, RLP6, receptor like protein 6 |
0.682 |
0.041 |
| 204 |
262164_at |
AT1G78070
|
[Transducin/WD40 repeat-like superfamily protein] |
0.680 |
-0.192 |
| 205 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
0.679 |
-0.207 |
| 206 |
250493_at |
AT5G09800
|
[ARM repeat superfamily protein] |
0.678 |
-0.071 |
| 207 |
260357_at |
AT1G69260
|
AFP1, ABI five binding protein |
0.678 |
-0.219 |
| 208 |
247030_at |
AT5G67210
|
IRX15-L, IRX15-LIKE |
0.676 |
-0.021 |
| 209 |
260205_at |
AT1G70700
|
JAZ9, JASMONATE-ZIM-DOMAIN PROTEIN 9, TIFY7 |
0.676 |
-0.072 |
| 210 |
258741_at |
AT3G05790
|
LON4, lon protease 4 |
0.675 |
0.033 |
| 211 |
255250_at |
AT4G05100
|
AtMYB74, myb domain protein 74, MYB74, myb domain protein 74 |
0.675 |
-0.349 |
| 212 |
259015_at |
AT3G07350
|
unknown |
0.674 |
-0.167 |
| 213 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.673 |
-0.145 |
| 214 |
258682_at |
AT3G08720
|
ATPK19, Arabidopsis thaliana protein kinase 19, ATPK2, ARABIDOPSIS THALIANA PROTEIN KINASE 2, ATS6K2, ARABIDOPSIS THALIANA SERINE/THREONINE PROTEIN KINASE 2, S6K2, serine/threonine protein kinase 2 |
0.671 |
0.008 |
| 215 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
0.669 |
-0.483 |
| 216 |
266413_at |
AT2G38740
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.668 |
-0.034 |
| 217 |
265216_at |
AT1G05100
|
MAPKKK18, mitogen-activated protein kinase kinase kinase 18 |
0.668 |
-0.184 |
| 218 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
0.668 |
-0.085 |
| 219 |
262103_at |
AT1G02940
|
ATGSTF5, GLUTATHIONE S-TRANSFERASE (CLASS PHI) 5, GSTF5, glutathione S-transferase (class phi) 5 |
0.667 |
0.042 |
| 220 |
256627_at |
AT3G19970
|
[alpha/beta-Hydrolases superfamily protein] |
0.662 |
-0.203 |
| 221 |
261443_at |
AT1G28480
|
GRX480, roxy19 |
0.661 |
-0.085 |
| 222 |
247041_at |
AT5G67180
|
TOE3, target of early activation tagged (EAT) 3 |
0.658 |
0.007 |
| 223 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.657 |
-0.197 |
| 224 |
247531_at |
AT5G61550
|
[U-box domain-containing protein kinase family protein] |
0.657 |
0.001 |
| 225 |
248392_at |
AT5G52050
|
[MATE efflux family protein] |
0.656 |
-0.120 |
| 226 |
260716_at |
AT1G48130
|
ATPER1, 1-cysteine peroxiredoxin 1, PER1, 1-cysteine peroxiredoxin 1 |
0.656 |
0.463 |
| 227 |
257946_at |
AT3G21710
|
unknown |
0.655 |
-0.131 |
| 228 |
261065_at |
AT1G07500
|
SMR5, SIAMESE-RELATED 5 |
0.655 |
-0.014 |
| 229 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
0.655 |
-0.085 |
| 230 |
247463_at |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
0.654 |
0.111 |
| 231 |
264783_at |
AT1G08650
|
ATPPCK1, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 1, PPCK1, phosphoenolpyruvate carboxylase kinase 1 |
0.653 |
-0.214 |
| 232 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
0.651 |
0.482 |
| 233 |
261033_at |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
0.648 |
0.229 |
| 234 |
254096_at |
AT4G25150
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
0.647 |
-0.067 |
| 235 |
252888_at |
AT4G39210
|
APL3 |
0.647 |
-0.022 |
| 236 |
249971_at |
AT5G19110
|
[Eukaryotic aspartyl protease family protein] |
0.646 |
0.070 |
| 237 |
266757_at |
AT2G46940
|
unknown |
0.645 |
-0.094 |
| 238 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.645 |
0.039 |
| 239 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.644 |
-0.159 |
| 240 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
0.643 |
0.002 |
| 241 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.641 |
-0.372 |
| 242 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
0.639 |
0.020 |
| 243 |
264891_at |
AT1G23200
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.636 |
-0.389 |
| 244 |
267206_at |
AT2G30830
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.635 |
-0.165 |
| 245 |
252638_at |
AT3G44540
|
FAR4, fatty acid reductase 4 |
0.635 |
-0.218 |
| 246 |
265356_at |
AT2G16595
|
[Translocon-associated protein (TRAP), alpha subunit] |
0.634 |
-0.177 |
| 247 |
265632_at |
AT2G14290
|
unknown |
0.631 |
-0.035 |
| 248 |
246551_at |
AT5G15070
|
[Phosphoglycerate mutase-like family protein] |
0.629 |
0.003 |
| 249 |
256285_at |
AT3G12510
|
[MADS-box family protein] |
0.627 |
-0.201 |
| 250 |
246251_at |
AT4G37220
|
[Cold acclimation protein WCOR413 family] |
0.626 |
-0.191 |
| 251 |
253643_at |
AT4G29780
|
unknown |
0.625 |
0.153 |
| 252 |
246386_at |
AT1G77410
|
BGAL16, beta-galactosidase 16 |
0.623 |
-0.041 |
| 253 |
256114_at |
AT1G16850
|
unknown |
0.623 |
-0.481 |
| 254 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
0.623 |
-0.406 |
| 255 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.621 |
-0.084 |
| 256 |
257217_at |
AT3G14940
|
ATPPC3, phosphoenolpyruvate carboxylase 3, PPC3, phosphoenolpyruvate carboxylase 3 |
0.620 |
-0.023 |
| 257 |
247243_at |
AT5G64700
|
UMAMIT21, Usually multiple acids move in and out Transporters 21 |
0.619 |
-0.193 |
| 258 |
251060_at |
AT5G01820
|
ATCIPK14, ATSR1, serine/threonine protein kinase 1, CIPK14, CBL-INTERACTING PROTEIN KINASE 14, PKS24, SOS2-like protein kinase 24, SnRK3.15, SNF1-RELATED PROTEIN KINASE 3.15, SR1, serine/threonine protein kinase 1 |
0.618 |
0.069 |
| 259 |
261636_at |
AT1G50110
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
0.617 |
0.158 |
| 260 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
0.617 |
0.046 |
| 261 |
267177_at |
AT2G37580
|
[RING/U-box superfamily protein] |
0.615 |
-0.117 |
| 262 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.614 |
-0.385 |
| 263 |
267497_at |
AT2G30540
|
[Thioredoxin superfamily protein] |
0.614 |
0.013 |
| 264 |
245889_at |
AT5G09480
|
[hydroxyproline-rich glycoprotein family protein] |
0.614 |
-0.249 |
| 265 |
251961_at |
AT3G53620
|
AtPPa4, pyrophosphorylase 4, PPa4, pyrophosphorylase 4 |
0.613 |
-0.169 |
| 266 |
266456_at |
AT2G22770
|
NAI1 |
0.612 |
0.024 |
| 267 |
258697_at |
AT3G09660
|
AtMCM8, MCM8, minichromosome maintenance 8 |
0.612 |
0.102 |
| 268 |
259705_at |
AT1G77450
|
anac032, NAC domain containing protein 32, NAC032, NAC domain containing protein 32 |
0.611 |
-0.312 |
| 269 |
262366_at |
AT1G72890
|
[Disease resistance protein (TIR-NBS class)] |
0.610 |
0.070 |
| 270 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
0.610 |
-0.004 |
| 271 |
248183_at |
AT5G54040
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.609 |
-0.160 |
| 272 |
266109_at |
AT2G37890
|
[Mitochondrial substrate carrier family protein] |
0.607 |
-0.057 |
| 273 |
266761_at |
AT2G47130
|
AtSDR3, SDR3, short-chain dehydrogenase/reductase 2 |
0.606 |
-0.070 |
| 274 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
0.606 |
-0.101 |
| 275 |
247359_at |
AT5G63560
|
FACT, FATTY ALCOHOL:CAFFEOYL-CoA CAFFEOYL TRANSFERASE |
0.605 |
-0.211 |
| 276 |
255587_at |
AT4G01480
|
AtPPa5, pyrophosphorylase 5, PPa5, pyrophosphorylase 5 |
0.604 |
0.018 |
| 277 |
250455_at |
AT5G09980
|
PROPEP4, elicitor peptide 4 precursor |
0.603 |
-0.038 |
| 278 |
265682_at |
AT2G24390
|
[AIG2-like (avirulence induced gene) family protein] |
0.603 |
0.097 |
| 279 |
249626_at |
AT5G37540
|
[Eukaryotic aspartyl protease family protein] |
0.601 |
-0.393 |
| 280 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
0.600 |
-0.216 |
| 281 |
251200_at |
AT3G63010
|
ATGID1B, GID1B, GA INSENSITIVE DWARF1B |
0.600 |
-0.163 |
| 282 |
251644_at |
AT3G57540
|
[Remorin family protein] |
0.599 |
-0.367 |
| 283 |
266092_at |
AT2G37880
|
unknown |
0.597 |
-0.039 |
| 284 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.597 |
-0.359 |
| 285 |
258901_at |
AT3G05640
|
[Protein phosphatase 2C family protein] |
0.597 |
-0.458 |
| 286 |
265913_at |
AT2G25625
|
unknown |
0.595 |
-0.285 |
| 287 |
255924_at |
AT1G22170
|
[Phosphoglycerate mutase family protein] |
0.593 |
0.066 |
| 288 |
255243_at |
AT4G05590
|
unknown |
0.593 |
0.059 |
| 289 |
259481_at |
AT1G18970
|
GLP4, germin-like protein 4 |
0.591 |
0.072 |
| 290 |
251066_at |
AT5G01880
|
DAFL2, DAF-Like gene 2 |
0.590 |
-0.301 |
| 291 |
250688_at |
AT5G06510
|
NF-YA10, nuclear factor Y, subunit A10 |
0.589 |
0.081 |
| 292 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.588 |
-0.239 |
| 293 |
246098_at |
AT5G20400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.583 |
-0.174 |
| 294 |
261785_at |
AT1G08230
|
[Transmembrane amino acid transporter family protein] |
0.583 |
-0.188 |
| 295 |
266690_at |
AT2G19900
|
ATNADP-ME1, Arabidopsis thaliana NADP-malic enzyme 1, NADP-ME1, NADP-malic enzyme 1 |
0.582 |
-0.166 |
| 296 |
257824_at |
AT3G25290
|
[Auxin-responsive family protein] |
0.582 |
0.089 |
| 297 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
0.582 |
-0.224 |
| 298 |
260386_at |
AT1G74010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.581 |
-0.133 |
| 299 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
0.580 |
-0.163 |
| 300 |
252114_at |
AT3G51450
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.579 |
0.033 |
| 301 |
251677_at |
AT3G56980
|
BHLH039, bHLH39, basic helix-loop-helix 39, ORG3, OBP3-RESPONSIVE GENE 3 |
-1.166 |
0.671 |
| 302 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-1.155 |
0.571 |
| 303 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-1.139 |
0.402 |
| 304 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-1.120 |
1.014 |
| 305 |
247573_at |
AT5G61160
|
AACT1, anthocyanin 5-aromatic acyltransferase 1 |
-1.077 |
-0.019 |
| 306 |
245692_at |
AT5G04150
|
BHLH101 |
-0.968 |
0.569 |
| 307 |
246275_at |
AT4G36540
|
BEE2, BR enhanced expression 2 |
-0.920 |
0.567 |
| 308 |
246273_at |
AT4G36700
|
[RmlC-like cupins superfamily protein] |
-0.904 |
0.210 |
| 309 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-0.851 |
0.418 |
| 310 |
265342_at |
AT2G18300
|
HBI1, HOMOLOG OF BEE2 INTERACTING WITH IBH 1 |
-0.849 |
0.941 |
| 311 |
246487_at |
AT5G16030
|
unknown |
-0.842 |
0.242 |
| 312 |
254931_at |
AT4G11460
|
CRK30, cysteine-rich RLK (RECEPTOR-like protein kinase) 30 |
-0.820 |
0.004 |
| 313 |
249378_at |
AT5G40450
|
unknown |
-0.813 |
-0.157 |
| 314 |
264507_at |
AT1G09415
|
NIMIN-3, NIM1-interacting 3 |
-0.786 |
0.013 |
| 315 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
-0.782 |
-1.119 |
| 316 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
-0.779 |
0.012 |
| 317 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
-0.778 |
0.205 |
| 318 |
254550_at |
AT4G19690
|
ATIRT1, ARABIDOPSIS IRON-REGULATED TRANSPORTER 1, IRT1, iron-regulated transporter 1 |
-0.771 |
-0.463 |
| 319 |
261684_at |
AT1G47400
|
unknown |
-0.762 |
0.808 |
| 320 |
259813_at |
AT1G49860
|
ATGSTF14, glutathione S-transferase (class phi) 14, GSTF14, glutathione S-transferase (class phi) 14 |
-0.748 |
-0.473 |
| 321 |
265116_at |
AT1G62480
|
[Vacuolar calcium-binding protein-related] |
-0.745 |
-0.012 |
| 322 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
-0.742 |
-0.203 |
| 323 |
254828_at |
AT4G12550
|
AIR1, Auxin-Induced in Root cultures 1 |
-0.715 |
0.272 |
| 324 |
259172_at |
AT3G03630
|
CS26, cysteine synthase 26 |
-0.704 |
0.007 |
| 325 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
-0.695 |
0.464 |
| 326 |
254233_at |
AT4G23800
|
3xHMG-box2, 3xHigh Mobility Group-box2 |
-0.661 |
0.393 |
| 327 |
256332_at |
AT1G76890
|
AT-GT2, GT2 |
-0.656 |
-0.091 |
| 328 |
249271_at |
AT5G41790
|
CIP1, COP1-interactive protein 1 |
-0.646 |
-0.011 |
| 329 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.643 |
0.447 |
| 330 |
258100_at |
AT3G23550
|
[MATE efflux family protein] |
-0.642 |
-0.111 |
| 331 |
264751_at |
AT1G23020
|
ATFRO3, FERRIC REDUCTION OXIDASE 3, FRO3, ferric reduction oxidase 3 |
-0.640 |
0.116 |
| 332 |
266957_at |
AT2G34640
|
HMR, HEMERA, PTAC12, plastid transcriptionally active 12, TAC12 |
-0.639 |
0.260 |
| 333 |
260343_at |
AT1G69200
|
FLN2, fructokinase-like 2 |
-0.628 |
0.228 |
| 334 |
247942_at |
AT5G57120
|
unknown |
-0.625 |
0.008 |
| 335 |
245925_at |
AT5G28770
|
AtbZIP63, Arabidopsis thaliana basic leucine zipper 63, BZO2H3 |
-0.623 |
0.081 |
| 336 |
260824_at |
AT1G06720
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.617 |
0.097 |
| 337 |
257759_at |
AT3G23070
|
ATCFM3A, CFM3A, CRM family member 3A |
-0.616 |
0.407 |
| 338 |
253278_at |
AT4G34220
|
[Leucine-rich repeat protein kinase family protein] |
-0.609 |
0.417 |
| 339 |
247032_at |
AT5G67240
|
SDN3, small RNA degrading nuclease 3 |
-0.606 |
-0.034 |
| 340 |
256675_at |
AT3G52170
|
[DNA binding] |
-0.598 |
0.425 |
| 341 |
261330_at |
AT1G44900
|
ATMCM2, MCM2, MINICHROMOSOME MAINTENANCE 2 |
-0.598 |
0.328 |
| 342 |
255774_at |
AT1G18620
|
TRM3, TON1 Recruiting Motif 3 |
-0.586 |
0.122 |
| 343 |
252362_at |
AT3G48500
|
PDE312, PIGMENT DEFECTIVE 312, PTAC10, PLASTID TRANSCRIPTIONALLY ACTIVE 10, TAC10 |
-0.578 |
0.071 |
| 344 |
255460_at |
AT4G02800
|
unknown |
-0.577 |
0.366 |
| 345 |
250828_at |
AT5G05250
|
unknown |
-0.567 |
0.165 |
| 346 |
260506_at |
AT1G47210
|
CYCA3;2, cyclin-dependent protein kinase 3;2 |
-0.566 |
0.164 |
| 347 |
257636_at |
AT3G26200
|
CYP71B22, cytochrome P450, family 71, subfamily B, polypeptide 22 |
-0.560 |
0.160 |
| 348 |
252904_at |
AT4G39620
|
ATPPR5, A. THALIANA PENTATRICOPEPTIDE REPEAT 5, EMB2453, EMBRYO DEFECTIVE 2453 |
-0.560 |
0.182 |
| 349 |
251800_at |
AT3G55510
|
RBL, REBELOTE |
-0.558 |
0.269 |
| 350 |
252089_at |
AT3G52110
|
unknown |
-0.553 |
-0.212 |
| 351 |
261660_at |
AT1G18370
|
ATNACK1, ARABIDOPSIS NPK1-ACTIVATING KINESIN 1, HIK, HINKEL, NACK1, NPK1-ACTIVATING KINESIN 1 |
-0.552 |
0.433 |
| 352 |
246303_at |
AT3G51870
|
[Mitochondrial substrate carrier family protein] |
-0.551 |
0.076 |
| 353 |
250560_at |
AT5G08020
|
ATRPA70B, ARABIDOPSIS THALIANA RPA70-KDA SUBUNIT B, RPA70B, RPA70-kDa subunit B |
-0.551 |
0.351 |
| 354 |
247606_at |
AT5G61000
|
ATRPA70D, RPA70D |
-0.549 |
0.346 |
| 355 |
254427_at |
AT4G21190
|
emb1417, embryo defective 1417 |
-0.548 |
0.134 |
| 356 |
265960_at |
AT2G37470
|
[Histone superfamily protein] |
-0.547 |
-0.023 |
| 357 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.542 |
0.581 |
| 358 |
248094_at |
AT5G55220
|
[trigger factor type chaperone family protein] |
-0.541 |
0.064 |
| 359 |
246415_at |
AT5G17160
|
unknown |
-0.541 |
0.288 |
| 360 |
260585_at |
AT2G43650
|
EMB2777, EMBRYO DEFECTIVE 2777 |
-0.541 |
0.150 |
| 361 |
254314_at |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
-0.540 |
0.464 |
| 362 |
255501_at |
AT4G02400
|
[U3 ribonucleoprotein (Utp) family protein] |
-0.537 |
0.098 |
| 363 |
249886_at |
AT5G22320
|
[Leucine-rich repeat (LRR) family protein] |
-0.526 |
0.301 |
| 364 |
247348_at |
AT5G63810
|
AtBGAL10, Arabidopsis thaliana beta-galactosidase 10, BGAL10, beta-galactosidase 10 |
-0.526 |
-0.155 |
| 365 |
254964_at |
AT4G11080
|
3xHMG-box1, 3xHigh Mobility Group-box1 |
-0.522 |
0.176 |
| 366 |
248656_at |
AT5G48460
|
[Actin binding Calponin homology (CH) domain-containing protein] |
-0.522 |
0.292 |
| 367 |
247284_at |
AT5G64410
|
ATOPT4, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 4, OPT4, oligopeptide transporter 4 |
-0.518 |
0.213 |
| 368 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
-0.513 |
0.732 |
| 369 |
266873_at |
AT2G44740
|
CYCP4;1, cyclin p4;1 |
-0.511 |
0.186 |
| 370 |
248709_at |
AT5G48470
|
PRDA1, PEP-Related Development Arrested 1 |
-0.511 |
-0.053 |
| 371 |
259839_at |
AT1G52190
|
AtNPF1.2, NPF1.2, NRT1/ PTR family 1.2, NRT1.11, nitrate transporter 1.11 |
-0.510 |
0.624 |
| 372 |
245426_at |
AT4G17540
|
unknown |
-0.509 |
0.387 |
| 373 |
255236_at |
AT4G05520
|
ATEHD2, EPS15 homology domain 2, EHD2, EPS15 homology domain 2 |
-0.507 |
0.143 |
| 374 |
254079_at |
AT4G25730
|
[FtsJ-like methyltransferase family protein] |
-0.504 |
0.105 |
| 375 |
250823_at |
AT5G05180
|
unknown |
-0.504 |
0.244 |
| 376 |
255128_at |
AT4G08310
|
unknown |
-0.500 |
0.044 |
| 377 |
246002_at |
AT5G20740
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.500 |
0.615 |
| 378 |
265464_at |
AT2G37080
|
RIP2, ROP interactive partner 2 |
-0.498 |
0.224 |
| 379 |
263485_at |
AT2G29890
|
ATVLN1, VLN1, villin 1 |
-0.497 |
0.138 |
| 380 |
254251_at |
AT4G23300
|
CRK22, cysteine-rich RLK (RECEPTOR-like protein kinase) 22 |
-0.495 |
0.446 |
| 381 |
265097_at |
AT1G04020
|
ATBARD1, BARD1, breast cancer associated RING 1, ROW1 |
-0.494 |
0.250 |
| 382 |
254572_at |
AT4G19380
|
[Long-chain fatty alcohol dehydrogenase family protein] |
-0.493 |
0.209 |
| 383 |
263677_at |
AT1G04520
|
PDLP2, plasmodesmata-located protein 2 |
-0.493 |
0.295 |
| 384 |
247235_at |
AT5G64580
|
EMB3144, EMBRYO DEFECTIVE 3144 |
-0.492 |
0.163 |
| 385 |
252361_at |
AT3G48490
|
unknown |
-0.489 |
0.274 |
| 386 |
248891_at |
AT5G46280
|
MCM3, MINICHROMOSOME MAINTENANCE 3 |
-0.488 |
0.400 |
| 387 |
251952_at |
AT3G53650
|
[Histone superfamily protein] |
-0.488 |
-0.097 |
| 388 |
252215_at |
AT3G50240
|
KICP-02 |
-0.488 |
-0.089 |
| 389 |
260799_at |
AT1G78270
|
AtUGT85A4, UDP-glucosyl transferase 85A4, UGT85A4, UDP-glucosyl transferase 85A4 |
-0.488 |
-0.007 |
| 390 |
246557_at |
AT5G15510
|
[TPX2 (targeting protein for Xklp2) protein family] |
-0.488 |
0.066 |
| 391 |
251935_at |
AT3G54090
|
FLN1, fructokinase-like 1 |
-0.488 |
0.057 |
| 392 |
265326_at |
AT2G18220
|
[Noc2p family] |
-0.487 |
0.142 |
| 393 |
260031_at |
AT1G68790
|
CRWN3, CROWDED NUCLEI 3, LINC3, LITTLE NUCLEI3 |
-0.486 |
0.169 |
| 394 |
252548_at |
AT3G45850
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.485 |
0.141 |
| 395 |
257062_at |
AT3G18290
|
BTS, BRUTUS, EMB2454, embryo defective 2454 |
-0.485 |
-0.038 |
| 396 |
264529_at |
AT1G30820
|
[CTP synthase family protein] |
-0.481 |
-0.298 |
| 397 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
-0.479 |
0.265 |
| 398 |
253421_at |
AT4G32340
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.478 |
0.018 |
| 399 |
264963_at |
AT1G60600
|
ABC4, ABERRANT CHLOROPLAST DEVELOPMENT 4 |
-0.476 |
0.270 |
| 400 |
261318_at |
AT1G53035
|
unknown |
-0.476 |
-0.240 |
| 401 |
267309_at |
AT2G19385
|
[zinc ion binding] |
-0.474 |
0.102 |
| 402 |
250461_at |
AT5G10010
|
HIT4, HEAT INTOLERANT 4 |
-0.473 |
0.059 |
| 403 |
249718_at |
AT5G35740
|
[Carbohydrate-binding X8 domain superfamily protein] |
-0.473 |
0.168 |
| 404 |
259278_at |
AT3G01160
|
unknown |
-0.473 |
0.088 |
| 405 |
259466_at |
AT1G19050
|
ARR7, response regulator 7 |
-0.472 |
0.144 |
| 406 |
249836_at |
AT5G23420
|
HMGB6, high-mobility group box 6 |
-0.471 |
0.532 |
| 407 |
254466_at |
AT4G20430
|
[Subtilase family protein] |
-0.470 |
0.216 |
| 408 |
263220_at |
AT1G30610
|
EMB2279, EMBRYO DEFECTIVE 2279, EMB88, EMBRYO DEFECTIVE 88 |
-0.469 |
0.209 |
| 409 |
247425_at |
AT5G62550
|
unknown |
-0.468 |
0.325 |
| 410 |
246269_at |
AT4G37110
|
[Zinc-finger domain of monoamine-oxidase A repressor R1] |
-0.468 |
0.143 |
| 411 |
254043_at |
AT4G25990
|
CIL |
-0.468 |
-0.330 |
| 412 |
266903_at |
AT2G34570
|
MEE21, maternal effect embryo arrest 21 |
-0.465 |
0.143 |
| 413 |
264114_at |
AT2G31270
|
ATCDT1A, ARABIDOPSIS HOMOLOG OF YEAST CDT1 A, CDT1, ARABIDOPSIS HOMOLOG OF YEAST CDT1, CDT1A, homolog of yeast CDT1 A |
-0.463 |
0.391 |
| 414 |
253975_at |
AT4G26600
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.461 |
0.276 |
| 415 |
260292_at |
AT1G63680
|
APG13, ALBINO OR PALE-GREEN 13, ATMURE, MURE, PDE316, PIGMENT DEFECTIVE EMBRYO 316 |
-0.460 |
0.128 |
| 416 |
258965_at |
AT3G10530
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.459 |
0.192 |
| 417 |
251759_at |
AT3G55630
|
ATDFD, DHFS-FPGS homolog D, DFD, DHFS-FPGS homolog D, FPGS3, folylpolyglutamate synthetase 3 |
-0.458 |
0.136 |
| 418 |
258545_at |
AT3G07050
|
NSN1, nucleostemin-like 1 |
-0.458 |
0.182 |
| 419 |
248413_at |
AT5G51600
|
ATMAP65-3, ARABIDOPSIS THALIANA MICROTUBULE-ASSOCIATED PROTEIN 65-3, MAP65-3, MICROTUBULE-ASSOCIATED PROTEIN 65-3, PLE, PLEIADE |
-0.457 |
0.260 |
| 420 |
249755_at |
AT5G24580
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.456 |
0.120 |
| 421 |
245876_at |
AT1G26230
|
Cpn60beta4, chaperonin-60beta4 |
-0.456 |
0.043 |
| 422 |
245411_at |
AT4G17240
|
unknown |
-0.455 |
-0.239 |
| 423 |
248895_at |
AT5G46330
|
FLS2, FLAGELLIN-SENSITIVE 2 |
-0.454 |
0.292 |
| 424 |
265042_at |
AT1G04040
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.452 |
-0.025 |
| 425 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
-0.452 |
-0.033 |
| 426 |
262207_at |
AT1G74850
|
PDE343, PIGMENT DEFECTIVE 343, PTAC2, plastid transcriptionally active 2 |
-0.451 |
0.008 |
| 427 |
249129_at |
AT5G43080
|
CYCA3;1, Cyclin A3;1 |
-0.450 |
0.268 |
| 428 |
264471_at |
AT1G67120
|
AtMDN1, MDN1, MIDASIN 1 |
-0.450 |
0.099 |
| 429 |
265085_at |
AT1G03780
|
AtTPX2, TPX2, targeting protein for XKLP2 |
-0.449 |
0.219 |
| 430 |
251852_at |
AT3G54750
|
unknown |
-0.445 |
0.155 |
| 431 |
262081_at |
AT1G59540
|
ZCF125 |
-0.445 |
0.222 |
| 432 |
251193_at |
AT3G62910
|
APG3, ALBINO AND PALE GREEN, AtcpRF1, Arabidopsis thaliana chloroplast RF1, cpRF1, chloroplast ribosome release factor 1 |
-0.444 |
-0.020 |
| 433 |
261761_at |
AT1G15660
|
CENP-C, CENP-C HOMOLOGUE, CENP-C, centromere protein C |
-0.444 |
-0.019 |
| 434 |
259303_at |
AT3G05130
|
unknown |
-0.443 |
0.143 |
| 435 |
264071_at |
AT2G27920
|
SCPL51, serine carboxypeptidase-like 51 |
-0.443 |
0.078 |
| 436 |
252077_at |
AT3G51720
|
unknown |
-0.442 |
0.085 |
| 437 |
254076_at |
AT4G25340
|
ATFKBP53, FKBP53, FK506 BINDING PROTEIN 53 |
-0.442 |
0.204 |
| 438 |
259928_at |
AT1G34380
|
[5'-3' exonuclease family protein] |
-0.441 |
-0.002 |
| 439 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
-0.441 |
0.016 |
| 440 |
253635_at |
AT4G30620
|
[Uncharacterised BCR, YbaB family COG0718] |
-0.440 |
0.096 |
| 441 |
248402_at |
AT5G52100
|
CRR1, chlororespiration reduction 1 |
-0.440 |
0.009 |
| 442 |
266616_at |
AT2G29680
|
ATCDC6, CDC6, cell division control 6 |
-0.439 |
0.241 |
| 443 |
257714_at |
AT3G27360
|
H3.1, histone 3.1 |
-0.437 |
0.101 |
| 444 |
247028_at |
AT5G67100
|
ICU2, INCURVATA2 |
-0.437 |
0.210 |
| 445 |
264096_at |
AT1G78995
|
unknown |
-0.435 |
0.318 |
| 446 |
263802_at |
AT2G40430
|
unknown |
-0.435 |
0.038 |
| 447 |
267578_at |
AT2G30695
|
unknown |
-0.434 |
0.170 |
| 448 |
248057_at |
AT5G55520
|
unknown |
-0.432 |
0.260 |
| 449 |
250255_at |
AT5G13730
|
SIG4, sigma factor 4, SIGD |
-0.432 |
0.061 |
| 450 |
262377_at |
AT1G73110
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.431 |
0.152 |
| 451 |
252034_at |
AT3G52040
|
unknown |
-0.431 |
0.126 |
| 452 |
259311_at |
AT3G05060
|
[NOP56-like pre RNA processing ribonucleoprotein] |
-0.428 |
0.113 |
| 453 |
263130_at |
AT1G78650
|
POLD3 |
-0.427 |
0.334 |
| 454 |
263558_at |
AT2G16380
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.427 |
0.010 |
| 455 |
263864_at |
AT2G04530
|
CPZ, TRZ2, TRNASE Z 2 |
-0.427 |
0.054 |
| 456 |
252149_at |
AT3G51290
|
APSR1, Altered Phosphate Starvation Response 1 |
-0.427 |
0.257 |
| 457 |
247098_at |
AT5G66470
|
[RNA binding] |
-0.425 |
0.142 |
| 458 |
249977_at |
AT5G18820
|
Cpn60alpha2, chaperonin-60alpha2, EMB3007, embryo defective 3007 |
-0.425 |
0.145 |
| 459 |
250825_at |
AT5G05210
|
[Surfeit locus protein 6] |
-0.424 |
-0.055 |
| 460 |
248177_at |
AT5G54630
|
[zinc finger protein-related] |
-0.424 |
0.410 |
| 461 |
254783_at |
AT4G12830
|
[alpha/beta-Hydrolases superfamily protein] |
-0.423 |
0.258 |
| 462 |
252691_at |
AT3G44050
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.423 |
0.141 |
| 463 |
266489_at |
AT2G35190
|
ATNPSN11, NPSN11, novel plant snare 11, NSPN11 |
-0.423 |
0.073 |
| 464 |
263679_at |
AT1G59990
|
EMB3108, EMBRYO DEFECTIVE 3108, HS3, HEAVY SEED 3, RH22, RNA helicase 22 |
-0.422 |
0.041 |
| 465 |
263607_at |
AT2G16270
|
unknown |
-0.421 |
0.176 |
| 466 |
263106_at |
AT2G05160
|
[CCCH-type zinc fingerfamily protein with RNA-binding domain] |
-0.421 |
0.026 |
| 467 |
254133_at |
AT4G24810
|
[Protein kinase superfamily protein] |
-0.420 |
0.196 |
| 468 |
259367_at |
AT1G69070
|
unknown |
-0.420 |
0.150 |
| 469 |
256786_at |
AT3G13740
|
[Ribonuclease III family protein] |
-0.420 |
-0.065 |
| 470 |
263766_at |
AT2G21440
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.418 |
0.156 |
| 471 |
249916_at |
AT5G22880
|
H2B, HISTONE H2B, HTB2, histone B2 |
-0.418 |
0.052 |
| 472 |
249900_at |
AT5G22640
|
emb1211, embryo defective 1211, TIC100, TRANSLOCON AT THE INNER ENVELOPE MEMBRANE OF CHLOROPLASTS 100 |
-0.417 |
0.096 |
| 473 |
252147_at |
AT3G51270
|
[protein serine/threonine kinases] |
-0.417 |
0.064 |
| 474 |
245709_at |
AT5G04320
|
[Shugoshin C terminus] |
-0.417 |
0.154 |
| 475 |
263039_at |
AT1G23280
|
[MAK16 protein-related] |
-0.416 |
0.119 |
| 476 |
249276_at |
AT5G41880
|
POLA3, POLA4 |
-0.415 |
0.256 |
| 477 |
263535_at |
AT2G24970
|
unknown |
-0.415 |
0.154 |
| 478 |
258630_at |
AT3G02820
|
[zinc knuckle (CCHC-type) family protein] |
-0.415 |
0.073 |
| 479 |
262766_at |
AT1G13160
|
[ARM repeat superfamily protein] |
-0.414 |
0.027 |
| 480 |
264872_at |
AT1G24260
|
AGL9, AGAMOUS-like 9, SEP3, SEPALLATA3 |
-0.414 |
0.201 |
| 481 |
261780_at |
AT1G76310
|
CYCB2;4, CYCLIN B2;4 |
-0.414 |
0.330 |
| 482 |
252410_at |
AT3G47450
|
ATNOA1, ATNOS1, NOA1, NO ASSOCIATED 1, NOS1, NITRIC OXIDE SYNTHASE 1, RIF1, RESISTANT TO INHIBITION WITH FOSMIDOMYCIN 1 |
-0.414 |
0.094 |
| 483 |
250100_at |
AT5G16570
|
GLN1;4, glutamine synthetase 1;4 |
-0.414 |
-0.063 |
| 484 |
255265_at |
AT4G05190
|
ATK5, kinesin 5 |
-0.414 |
0.215 |
| 485 |
259151_at |
AT3G10310
|
[P-loop nucleoside triphosphate hydrolases superfamily protein with CH (Calponin Homology) domain] |
-0.413 |
0.117 |
| 486 |
260948_at |
AT1G06100
|
[Fatty acid desaturase family protein] |
-0.411 |
-0.031 |
| 487 |
248344_at |
AT5G52280
|
[Myosin heavy chain-related protein] |
-0.411 |
0.017 |
| 488 |
254250_at |
AT4G23290
|
CRK21, cysteine-rich RLK (RECEPTOR-like protein kinase) 21 |
-0.410 |
0.255 |
| 489 |
257667_at |
AT3G20440
|
BE1, BRANCHING ENZYME 1, EMB2729, EMBRYO DEFECTIVE 2729 |
-0.407 |
0.071 |
| 490 |
261610_at |
AT1G49560
|
[Homeodomain-like superfamily protein] |
-0.406 |
-0.130 |
| 491 |
259344_at |
AT3G03710
|
PDE326, PIGMENT DEFECTIVE 326, PNP, POLYNUCLEOTIDE PHOSPHORYLASE, RIF10, resistant to inhibition with FSM 10 |
-0.406 |
0.025 |
| 492 |
253156_at |
AT4G35730
|
[Regulator of Vps4 activity in the MVB pathway protein] |
-0.406 |
0.032 |
| 493 |
261301_at |
AT1G48570
|
[zinc finger (Ran-binding) family protein] |
-0.406 |
0.247 |
| 494 |
253051_at |
AT4G37490
|
CYC1, CYCLIN 1, CYCB1, CYCB1;1, CYCLIN B1;1 |
-0.405 |
0.253 |
| 495 |
247603_at |
AT5G60930
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.404 |
0.119 |
| 496 |
253804_at |
AT4G28230
|
unknown |
-0.403 |
0.160 |
| 497 |
248493_at |
AT5G51100
|
FSD2, Fe superoxide dismutase 2 |
-0.403 |
0.039 |
| 498 |
255465_at |
AT4G02990
|
BSM, BELAYA SMERT, RUG2, RUGOSA 2 |
-0.403 |
0.245 |
| 499 |
248270_at |
AT5G53450
|
ORG1, OBP3-responsive gene 1 |
-0.403 |
0.036 |
| 500 |
265340_at |
AT2G18330
|
[AAA-type ATPase family protein] |
-0.402 |
0.234 |
| 501 |
261942_at |
AT1G22590
|
AGL87, AGAMOUS-like 87 |
-0.401 |
-0.014 |
| 502 |
251261_at |
AT3G62110
|
[Pectin lyase-like superfamily protein] |
-0.401 |
0.196 |
| 503 |
259822_at |
AT1G66230
|
AtMYB20, myb domain protein 20, MYB20, myb domain protein 20 |
-0.399 |
0.008 |
| 504 |
260815_at |
AT1G06950
|
ATTIC110, ARABIDOPSIS THALIANA TRANSLOCON AT THE INNER ENVELOPE MEMBRANE OF CHLOROPLASTS 110, TIC110, translocon at the inner envelope membrane of chloroplasts 110 |
-0.398 |
0.083 |
| 505 |
250711_at |
AT5G06110
|
[DnaJ domain ] |
-0.398 |
-0.065 |
| 506 |
257534_at |
AT3G09670
|
[Tudor/PWWP/MBT superfamily protein] |
-0.398 |
-0.147 |
| 507 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
-0.396 |
-0.040 |
| 508 |
258554_at |
AT3G06980
|
[DEA(D/H)-box RNA helicase family protein] |
-0.395 |
0.068 |
| 509 |
258138_at |
AT3G24495
|
ATMSH7, ARABIDOPSIS THALIANA MUTS HOMOLOG 7, MSH6-2, MUTS HOMOLOG 6-2, MSH7, MUTS homolog 7 |
-0.394 |
0.171 |
| 510 |
265274_at |
AT2G28450
|
[zinc finger (CCCH-type) family protein] |
-0.394 |
0.083 |
| 511 |
265336_at |
AT2G18290
|
APC10, anaphase promoting complex 10, AtAPC10, EMB2783, EMBRYO DEFECTIVE 2783 |
-0.394 |
0.207 |
| 512 |
258920_at |
AT3G10520
|
AHB2, haemoglobin 2, ARATH GLB2, ATGLB2, ARABIDOPSIS HEMOGLOBIN 2, GLB2, HEMOGLOBIN 2, HB2, haemoglobin 2, NSHB2, NON-SYMBIOTIC HAEMOGLOBIN 2 |
-0.393 |
0.285 |
| 513 |
259210_at |
AT3G09150
|
ATHY2, ARABIDOPSIS ELONGATED HYPOCOTYL 2, GUN3, GENOMES UNCOUPLED 3, HY2, ELONGATED HYPOCOTYL 2 |
-0.393 |
0.093 |
| 514 |
251244_at |
AT3G62060
|
[Pectinacetylesterase family protein] |
-0.393 |
0.256 |
| 515 |
259686_at |
AT1G63100
|
[GRAS family transcription factor] |
-0.392 |
0.072 |
| 516 |
250529_at |
AT5G08610
|
PDE340, PIGMENT DEFECTIVE 340 |
-0.392 |
0.108 |
| 517 |
266959_at |
AT2G07690
|
MCM5, MINICHROMOSOME MAINTENANCE 5 |
-0.390 |
0.275 |
| 518 |
250394_at |
AT5G10910
|
[mraW methylase family protein] |
-0.390 |
0.333 |
| 519 |
252148_at |
AT3G51280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.389 |
0.359 |
| 520 |
253817_at |
AT4G28310
|
unknown |
-0.389 |
0.306 |
| 521 |
249742_at |
AT5G24490
|
[30S ribosomal protein, putative] |
-0.387 |
-0.053 |
| 522 |
255637_at |
AT4G00750
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.387 |
-0.245 |
| 523 |
255874_at |
AT2G40550
|
ETG1, E2F target gene 1 |
-0.386 |
0.280 |
| 524 |
266668_at |
AT2G29760
|
OTP81, ORGANELLE TRANSCRIPT PROCESSING 81 |
-0.386 |
-0.057 |
| 525 |
256864_at |
AT3G23890
|
ATTOPII, TOPII, topoisomerase II |
-0.386 |
0.108 |
| 526 |
251371_at |
AT3G60360
|
EDA14, EMBRYO SAC DEVELOPMENT ARREST 14, UTP11, U3 SMALL NUCLEOLAR RNA-ASSOCIATED PROTEIN 11 |
-0.386 |
-0.025 |
| 527 |
245291_at |
AT4G16155
|
[dihydrolipoyl dehydrogenases] |
-0.384 |
0.073 |
| 528 |
258859_at |
AT3G02120
|
[hydroxyproline-rich glycoprotein family protein] |
-0.384 |
0.367 |
| 529 |
265588_at |
AT2G19970
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.383 |
-0.168 |
| 530 |
249528_at |
AT5G38720
|
unknown |
-0.383 |
0.147 |
| 531 |
246095_at |
AT5G19310
|
CHR23, chromatin remodeling 23 |
-0.383 |
-0.374 |
| 532 |
252765_at |
AT3G42800
|
unknown |
-0.381 |
0.064 |
| 533 |
257421_at |
AT1G12030
|
unknown |
-0.380 |
0.245 |
| 534 |
253403_at |
AT4G32830
|
AtAUR1, ataurora1, AUR1, ataurora1 |
-0.380 |
0.200 |
| 535 |
251969_at |
AT3G53130
|
CYP97C1, CYTOCHROME P450 97C1, LUT1, LUTEIN DEFICIENT 1 |
-0.380 |
0.142 |
| 536 |
255328_at |
AT4G04350
|
EMB2369, EMBRYO DEFECTIVE 2369 |
-0.379 |
0.061 |
| 537 |
245343_at |
AT4G15830
|
[ARM repeat superfamily protein] |
-0.379 |
0.439 |
| 538 |
264969_at |
AT1G67320
|
EMB2813, EMBRYO DEFECTIVE 2813 |
-0.379 |
0.128 |
| 539 |
255513_at |
AT4G02060
|
MCM7, PRL, PROLIFERA |
-0.378 |
0.302 |
| 540 |
246457_at |
AT5G16750
|
TOZ, TORMOZEMBRYO DEFECTIVE |
-0.378 |
-0.014 |
| 541 |
257005_at |
AT3G14190
|
CMR1, COPPER MODIFIED RESISTANCE 1 |
-0.378 |
0.223 |
| 542 |
261577_at |
AT1G01080
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.378 |
0.278 |
| 543 |
261762_at |
AT1G15510
|
ATECB2, ARABIDOPSIS EARLY CHLOROPLAST BIOGENESIS2, ECB2, EARLY CHLOROPLAST BIOGENESIS2, VAC1, VANILLA CREAM 1 |
-0.377 |
0.165 |
| 544 |
247056_at |
AT5G66750
|
ATDDM1, CHA1, CHR01, CHROMATIN REMODELING 1, CHR1, chromatin remodeling 1, DDM1, DECREASED DNA METHYLATION 1, SOM1, SOMNIFEROUS 1, SOM4 |
-0.377 |
0.185 |
| 545 |
266687_at |
AT2G19670
|
ATPRMT1A, ARABIDOPSIS THALIANA PROTEIN ARGININE METHYLTRANSFERASE 1A, PRMT1A, protein arginine methyltransferase 1A |
-0.377 |
0.193 |
| 546 |
249008_at |
AT5G44680
|
[DNA glycosylase superfamily protein] |
-0.376 |
0.609 |
| 547 |
265695_at |
AT2G24490
|
ATRPA2, ATRPA32A, ROR1, SUPPRESSOR OF ROS1, RPA2, replicon protein A2, RPA32A |
-0.376 |
0.109 |
| 548 |
252785_at |
AT3G42660
|
[transducin family protein / WD-40 repeat family protein] |
-0.375 |
0.176 |
| 549 |
265991_at |
AT2G24120
|
PDE319, PIGMENT DEFECTIVE 319, SCA3, SCABRA 3 |
-0.375 |
0.054 |
| 550 |
250418_at |
AT5G11240
|
[transducin family protein / WD-40 repeat family protein] |
-0.374 |
0.189 |
| 551 |
257201_at |
AT3G23740
|
unknown |
-0.374 |
0.140 |
| 552 |
250649_at |
AT5G06690
|
WCRKC1, WCRKC thioredoxin 1 |
-0.373 |
-0.175 |
| 553 |
258782_at |
AT3G11750
|
FOLB1 |
-0.373 |
0.136 |
| 554 |
262706_at |
AT1G16280
|
AtRH36, Arabidopsis thaliana RNA helicase 36, RH36, RNA helicase 36, SWA3, SLOW WALKER 3 |
-0.373 |
0.191 |
| 555 |
255053_at |
AT4G09730
|
RH39, RH39 |
-0.373 |
0.143 |
| 556 |
264121_at |
AT1G02280
|
ATTOC33, PPI1, PLASTID PROTEIN IMPORT 1, TOC33, translocon at the outer envelope membrane of chloroplasts 33 |
-0.372 |
0.047 |
| 557 |
253977_at |
AT4G26630
|
[DEK domain-containing chromatin associated protein] |
-0.372 |
0.100 |
| 558 |
250528_at |
AT5G08600
|
[U3 ribonucleoprotein (Utp) family protein] |
-0.372 |
0.117 |
| 559 |
262043_at |
AT1G80190
|
PSF1, partner of SLD five 1 |
-0.372 |
0.010 |
| 560 |
261460_at |
AT1G07880
|
ATMPK13 |
-0.372 |
0.169 |
| 561 |
258573_at |
AT3G04260
|
PDE324, PIGMENT DEFECTIVE 324, PTAC3, plastid transcriptionally active 3 |
-0.372 |
0.029 |
| 562 |
252625_at |
AT3G44750
|
ATHD2A, HD2A, HISTONE DEACETYLASE 2A, HDA3, histone deacetylase 3, HDT1 |
-0.371 |
0.168 |
| 563 |
246765_at |
AT5G27330
|
[Prefoldin chaperone subunit family protein] |
-0.371 |
0.209 |
| 564 |
250679_at |
AT5G06550
|
JMJ22, Jumonji domain-containing protein 22 |
-0.371 |
0.140 |
| 565 |
247962_at |
AT5G56580
|
ANQ1, ARABIDOPSIS NQK1, ATMKK6, ARABIDOPSIS THALIANA MAP KINASE KINASE 6, MKK6, MAP kinase kinase 6 |
-0.371 |
0.171 |
| 566 |
261911_at |
AT1G80750
|
[Ribosomal protein L30/L7 family protein] |
-0.371 |
0.075 |
| 567 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
-0.371 |
-0.363 |
| 568 |
263612_at |
AT2G16440
|
MCM4, MINICHROMOSOME MAINTENANCE 4 |
-0.370 |
0.240 |
| 569 |
249366_at |
AT5G40610
|
GPDHp, Glycerol-3-phosphate dehydrogenase plastidic |
-0.370 |
0.087 |
| 570 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
-0.370 |
0.755 |
| 571 |
264748_at |
AT1G70070
|
EMB25, EMBRYO DEFECTIVE 25, ISE2, INCREASED SIZE EXCLUSION LIMIT 2, PDE317, PIGMENT DEFECTIVE 317 |
-0.369 |
0.091 |
| 572 |
251355_at |
AT3G61100
|
[Putative endonuclease or glycosyl hydrolase] |
-0.369 |
0.267 |
| 573 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
-0.368 |
0.601 |
| 574 |
254221_at |
AT4G23820
|
[Pectin lyase-like superfamily protein] |
-0.368 |
0.265 |
| 575 |
253978_at |
AT4G26660
|
unknown |
-0.367 |
0.168 |
| 576 |
247046_at |
AT5G66540
|
unknown |
-0.367 |
0.075 |
| 577 |
254281_at |
AT4G22840
|
[Sodium Bile acid symporter family] |
-0.366 |
0.118 |
| 578 |
245117_at |
AT2G41560
|
ACA4, autoinhibited Ca(2+)-ATPase, isoform 4 |
-0.366 |
0.015 |
| 579 |
255278_at |
AT4G04940
|
[transducin family protein / WD-40 repeat family protein] |
-0.366 |
0.243 |
| 580 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
-0.365 |
0.294 |
| 581 |
259851_at |
AT1G72250
|
[Di-glucose binding protein with Kinesin motor domain] |
-0.364 |
0.189 |
| 582 |
256077_at |
AT1G18090
|
[5'-3' exonuclease family protein] |
-0.363 |
0.311 |
| 583 |
249538_at |
AT5G38840
|
[SMAD/FHA domain-containing protein ] |
-0.362 |
-0.014 |
| 584 |
262698_at |
AT1G75960
|
[AMP-dependent synthetase and ligase family protein] |
-0.362 |
0.187 |
| 585 |
266478_at |
AT2G31170
|
FIONA, FIONA, SYCO ARATH, cysteinyl t-RNA synthetase |
-0.361 |
0.161 |
| 586 |
256797_at |
AT3G18600
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.361 |
0.176 |
| 587 |
253887_at |
AT4G27730
|
ATOPT6, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 6, OPT6, oligopeptide transporter 1 |
-0.360 |
0.163 |
| 588 |
260431_at |
AT1G68190
|
BBX27, B-box domain protein 27 |
-0.360 |
-0.061 |
| 589 |
261611_at |
AT1G49730
|
[Protein kinase superfamily protein] |
-0.360 |
-0.100 |
| 590 |
267562_at |
AT2G39670
|
[Radical SAM superfamily protein] |
-0.359 |
0.072 |
| 591 |
262777_at |
AT1G13030
|
Atcoilin, COILIN, coilin |
-0.359 |
0.108 |
| 592 |
262453_at |
AT1G11240
|
unknown |
-0.358 |
-0.021 |
| 593 |
246919_at |
AT5G25460
|
DGR2, DUF642 L-GalL responsive gene 2 |
-0.358 |
0.250 |
| 594 |
266575_at |
AT2G24060
|
[Translation initiation factor 3 protein] |
-0.358 |
0.132 |
| 595 |
249993_at |
AT5G18570
|
ATOBGC, ATOBGL, OBG-like protein, CPSAR1, chloroplastic SAR1, EMB269, EMBRYO DEFECTIVE 269, EMB3138, EMBRYO DEFECTIVE 3138 |
-0.357 |
0.067 |
| 596 |
264802_at |
AT1G08560
|
ATSYP111, KN, KNOLLE, SYP111, syntaxin of plants 111 |
-0.357 |
0.197 |
| 597 |
253566_at |
AT4G31210
|
[DNA topoisomerase, type IA, core] |
-0.356 |
0.205 |
| 598 |
255856_at |
AT1G66940
|
[protein kinase-related] |
-0.355 |
0.363 |
| 599 |
256547_at |
AT3G14840
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.355 |
0.114 |
| 600 |
257207_at |
AT3G14900
|
EMB3120, EMBRYO DEFECTIVE 3120 |
-0.355 |
0.157 |