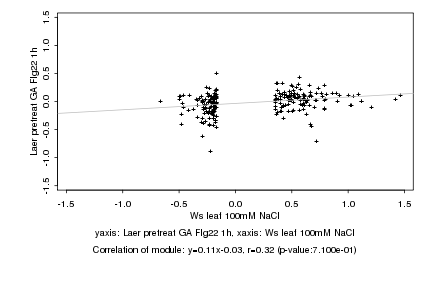
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Ws leaf 100mM NaCl |
Log2 signal ratio
Laer pretreat GA Flg22 1h |
| 1 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.461 |
0.121 |
| 2 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
1.417 |
0.041 |
| 3 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
1.204 |
-0.103 |
| 4 |
259286_at |
AT3G11480
|
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
1.117 |
0.014 |
| 5 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
1.114 |
0.015 |
| 6 |
264580_at |
AT1G05340
|
unknown |
1.085 |
0.138 |
| 7 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
1.045 |
0.103 |
| 8 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
1.026 |
-0.061 |
| 9 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
1.019 |
-0.065 |
| 10 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
1.000 |
0.115 |
| 11 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
0.918 |
0.108 |
| 12 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
0.901 |
0.007 |
| 13 |
258225_at |
AT3G15630
|
unknown |
0.891 |
0.147 |
| 14 |
260744_at |
AT1G15010
|
unknown |
0.858 |
0.154 |
| 15 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
0.805 |
0.136 |
| 16 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.792 |
0.053 |
| 17 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
0.788 |
-0.137 |
| 18 |
254234_at |
AT4G23680
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.785 |
-0.116 |
| 19 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.783 |
0.027 |
| 20 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
0.783 |
0.289 |
| 21 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.766 |
0.103 |
| 22 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
0.763 |
0.158 |
| 23 |
253628_at |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
0.730 |
0.238 |
| 24 |
251668_at |
AT3G57010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.716 |
0.031 |
| 25 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
0.716 |
0.148 |
| 26 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
0.715 |
-0.708 |
| 27 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.702 |
0.035 |
| 28 |
261658_at |
AT1G50040
|
unknown |
0.696 |
-0.095 |
| 29 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
0.671 |
-0.443 |
| 30 |
246929_at |
AT5G25210
|
unknown |
0.668 |
0.090 |
| 31 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.661 |
0.166 |
| 32 |
248611_at |
AT5G49520
|
ATWRKY48, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 48, WRKY48, WRKY DNA-binding protein 48 |
0.661 |
0.111 |
| 33 |
252487_at |
AT3G46660
|
UGT76E12, UDP-glucosyl transferase 76E12 |
0.659 |
-0.408 |
| 34 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
0.656 |
0.299 |
| 35 |
267147_at |
AT2G38240
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.651 |
0.095 |
| 36 |
253485_at |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
0.650 |
-0.056 |
| 37 |
247013_at |
AT5G67480
|
ATBT4, BT4, BTB and TAZ domain protein 4 |
0.639 |
0.052 |
| 38 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
0.635 |
0.027 |
| 39 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.630 |
-0.013 |
| 40 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
0.622 |
-0.224 |
| 41 |
265276_at |
AT2G28400
|
unknown |
0.615 |
0.023 |
| 42 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.612 |
0.108 |
| 43 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
0.608 |
0.103 |
| 44 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.606 |
-0.065 |
| 45 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.603 |
-0.021 |
| 46 |
267246_at |
AT2G30250
|
ATWRKY25, WRKY25, WRKY DNA-binding protein 25 |
0.602 |
0.135 |
| 47 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
0.600 |
0.052 |
| 48 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
0.599 |
-0.126 |
| 49 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.597 |
0.071 |
| 50 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
0.588 |
-0.066 |
| 51 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
0.576 |
0.215 |
| 52 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
0.575 |
0.031 |
| 53 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
0.572 |
-0.053 |
| 54 |
246253_at |
AT4G37260
|
ATMYB73, MYB73, myb domain protein 73 |
0.568 |
-0.147 |
| 55 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
0.562 |
0.433 |
| 56 |
254652_at |
AT4G18170
|
ATWRKY28, WRKY28, WRKY DNA-binding protein 28 |
0.560 |
0.100 |
| 57 |
264774_at |
AT1G22890
|
unknown |
0.558 |
0.312 |
| 58 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
0.557 |
0.075 |
| 59 |
253643_at |
AT4G29780
|
unknown |
0.550 |
0.070 |
| 60 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
0.546 |
0.129 |
| 61 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
0.540 |
0.264 |
| 62 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.528 |
0.073 |
| 63 |
259364_at |
AT1G13260
|
EDF4, ETHYLENE RESPONSE DNA BINDING FACTOR 4, RAV1, related to ABI3/VP1 1 |
0.520 |
0.086 |
| 64 |
266259_at |
AT2G27830
|
unknown |
0.516 |
0.146 |
| 65 |
254574_at |
AT4G19430
|
unknown |
0.515 |
-0.155 |
| 66 |
266658_at |
AT2G25735
|
unknown |
0.514 |
-0.014 |
| 67 |
251432_at |
AT3G59820
|
AtLETM1, LETM1, leucine zipper-EF-hand-containing transmembrane protein 1 |
0.512 |
0.200 |
| 68 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
0.511 |
0.110 |
| 69 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
0.506 |
-0.166 |
| 70 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
0.502 |
0.270 |
| 71 |
266316_at |
AT2G27080
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.497 |
0.125 |
| 72 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
0.497 |
0.147 |
| 73 |
258939_at |
AT3G10020
|
unknown |
0.495 |
0.047 |
| 74 |
256833_at |
AT3G22910
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.494 |
0.290 |
| 75 |
265147_at |
AT1G51380
|
[DEA(D/H)-box RNA helicase family protein] |
0.494 |
0.036 |
| 76 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
0.488 |
0.033 |
| 77 |
251928_at |
AT3G53980
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.487 |
0.094 |
| 78 |
245252_at |
AT4G17500
|
ATERF-1, ethylene responsive element binding factor 1, ERF-1, ethylene responsive element binding factor 1 |
0.478 |
0.092 |
| 79 |
250158_at |
AT5G15190
|
unknown |
0.478 |
0.108 |
| 80 |
251507_at |
AT3G59080
|
[Eukaryotic aspartyl protease family protein] |
0.474 |
0.189 |
| 81 |
261193_at |
AT1G32920
|
unknown |
0.472 |
0.030 |
| 82 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
0.470 |
-0.039 |
| 83 |
259729_at |
AT1G77640
|
[Integrase-type DNA-binding superfamily protein] |
0.466 |
-0.166 |
| 84 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
0.464 |
0.034 |
| 85 |
259911_at |
AT1G72680
|
ATCAD1, CINNAMYL ALCOHOL DEHYDROGENASE 1, CAD1, cinnamyl-alcohol dehydrogenase |
0.463 |
0.035 |
| 86 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
0.459 |
0.086 |
| 87 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.452 |
0.118 |
| 88 |
246200_at |
AT4G37240
|
unknown |
0.436 |
-0.064 |
| 89 |
247540_at |
AT5G61590
|
[Integrase-type DNA-binding superfamily protein] |
0.436 |
-0.059 |
| 90 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
0.431 |
0.131 |
| 91 |
263754_at |
AT2G21510
|
[DNAJ heat shock N-terminal domain-containing protein] |
0.429 |
0.170 |
| 92 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
0.427 |
0.088 |
| 93 |
252320_at |
AT3G48580
|
XTH11, xyloglucan endotransglucosylase/hydrolase 11 |
0.426 |
-0.289 |
| 94 |
257076_at |
AT3G19680
|
unknown |
0.423 |
-0.088 |
| 95 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
0.421 |
-0.069 |
| 96 |
264652_at |
AT1G08920
|
ESL1, ERD (early response to dehydration) six-like 1 |
0.416 |
0.330 |
| 97 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
0.412 |
0.130 |
| 98 |
261727_at |
AT1G76090
|
SMT3, sterol methyltransferase 3 |
0.406 |
-0.166 |
| 99 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
0.401 |
-0.099 |
| 100 |
262336_at |
AT1G64220
|
TOM7-2, translocase of outer membrane 7 kDa subunit 2 |
0.399 |
-0.033 |
| 101 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.396 |
0.103 |
| 102 |
263799_at |
AT2G24550
|
unknown |
0.393 |
0.052 |
| 103 |
252193_at |
AT3G50060
|
MYB77, myb domain protein 77 |
0.391 |
-0.168 |
| 104 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
0.389 |
0.144 |
| 105 |
263475_at |
AT2G31945
|
unknown |
0.385 |
0.156 |
| 106 |
251192_at |
AT3G62720
|
ATXT1, XT1, xylosyltransferase 1, XXT1, XYG XYLOSYLTRANSFERASE 1 |
0.376 |
0.052 |
| 107 |
260126_at |
AT1G36370
|
SHM7, serine hydroxymethyltransferase 7 |
0.373 |
0.201 |
| 108 |
261926_at |
AT1G22530
|
PATL2, PATELLIN 2 |
0.373 |
-0.190 |
| 109 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.367 |
0.335 |
| 110 |
261242_at |
AT1G32960
|
ATSBT3.3, SBT3.3 |
0.360 |
0.338 |
| 111 |
251221_at |
AT3G62550
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.359 |
-0.217 |
| 112 |
259979_at |
AT1G76600
|
unknown |
0.358 |
0.059 |
| 113 |
245041_at |
AT2G26530
|
AR781 |
0.356 |
0.051 |
| 114 |
264246_at |
AT1G60140
|
ATTPS10, trehalose phosphate synthase, TPS10, trehalose phosphate synthase, TPS10, TREHALOSE PHOSPHATE SYNTHASE 10 |
0.355 |
0.119 |
| 115 |
252305_at |
AT3G49240
|
emb1796, embryo defective 1796 |
0.354 |
-0.004 |
| 116 |
250365_at |
AT5G11410
|
[Protein kinase superfamily protein] |
0.353 |
-0.139 |
| 117 |
264186_at |
AT1G54570
|
PES1, phytyl ester synthase 1 |
0.351 |
-0.081 |
| 118 |
254580_at |
AT4G19390
|
[Uncharacterised protein family (UPF0114)] |
0.348 |
0.033 |
| 119 |
267028_at |
AT2G38470
|
ATWRKY33, WRKY DNA-BINDING PROTEIN 33, WRKY33, WRKY DNA-binding protein 33 |
0.347 |
0.052 |
| 120 |
254931_at |
AT4G11460
|
CRK30, cysteine-rich RLK (RECEPTOR-like protein kinase) 30 |
-0.665 |
0.018 |
| 121 |
252501_at |
AT3G46880
|
unknown |
-0.505 |
0.097 |
| 122 |
264083_at |
AT2G31230
|
ATERF15, ethylene-responsive element binding factor 15, ERF15, ethylene-responsive element binding factor 15 |
-0.505 |
0.048 |
| 123 |
261574_at |
AT1G01190
|
CYP78A8, cytochrome P450, family 78, subfamily A, polypeptide 8 |
-0.488 |
0.098 |
| 124 |
253940_at |
AT4G26950
|
unknown |
-0.482 |
-0.216 |
| 125 |
263133_at |
AT1G78450
|
[SOUL heme-binding family protein] |
-0.481 |
-0.405 |
| 126 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-0.477 |
-0.032 |
| 127 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.465 |
0.114 |
| 128 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
-0.462 |
-0.101 |
| 129 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
-0.425 |
-0.155 |
| 130 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.414 |
0.117 |
| 131 |
251402_at |
AT3G60290
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.381 |
-0.131 |
| 132 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
-0.349 |
0.047 |
| 133 |
258287_at |
AT3G15990
|
SULTR3;4, sulfate transporter 3;4 |
-0.341 |
0.031 |
| 134 |
250471_at |
AT5G10170
|
ATMIPS3, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 3, MIPS3, myo-inositol-1-phosphate synthase 3 |
-0.341 |
-0.057 |
| 135 |
262290_at |
AT1G70985
|
[hydroxyproline-rich glycoprotein family protein] |
-0.338 |
-0.267 |
| 136 |
250744_at |
AT5G05840
|
unknown |
-0.319 |
0.055 |
| 137 |
264998_at |
AT1G67330
|
unknown |
-0.310 |
-0.367 |
| 138 |
256780_at |
AT3G13640
|
ABCE1, ATP-binding cassette E1, ATRLI1, RNAse l inhibitor protein 1, RLI1, RNAse l inhibitor protein 1 |
-0.303 |
0.096 |
| 139 |
261068_at |
AT1G07450
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.301 |
0.016 |
| 140 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-0.300 |
-0.619 |
| 141 |
262120_at |
AT1G02950
|
ATGSTF4, glutathione S-transferase F4, GST31, GLUTATHIONE S-TRANSFERASE 31, GSTF4, glutathione S-transferase F4 |
-0.299 |
-0.015 |
| 142 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
-0.293 |
0.021 |
| 143 |
260805_at |
AT1G78320
|
ATGSTU23, glutathione S-transferase TAU 23, GSTU23, glutathione S-transferase TAU 23 |
-0.293 |
-0.290 |
| 144 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
-0.292 |
-0.381 |
| 145 |
263777_at |
AT2G46450
|
ATCNGC12, cyclic nucleotide-gated channel 12, CNGC12, cyclic nucleotide-gated channel 12 |
-0.291 |
-0.138 |
| 146 |
244983_at |
ATCG00790
|
RPL16, ribosomal protein L16 |
-0.290 |
-0.099 |
| 147 |
245090_at |
AT2G40900
|
UMAMIT11, Usually multiple acids move in and out Transporters 11 |
-0.288 |
0.008 |
| 148 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.281 |
-0.203 |
| 149 |
260059_at |
AT1G78090
|
ATTPPB, Arabidopsis thaliana trehalose-6-phosphate phosphatase B, TPPB, trehalose-6-phosphate phosphatase B |
-0.280 |
-0.050 |
| 150 |
254870_at |
AT4G11900
|
[S-locus lectin protein kinase family protein] |
-0.274 |
-0.348 |
| 151 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
-0.271 |
-0.094 |
| 152 |
265962_at |
AT2G37460
|
UMAMIT12, Usually multiple acids move in and out Transporters 12 |
-0.266 |
-0.111 |
| 153 |
254545_at |
AT4G19830
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.266 |
-0.112 |
| 154 |
257120_at |
AT3G20200
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.266 |
0.037 |
| 155 |
245049_at |
ATCG00050
|
RPS16, ribosomal protein S16 |
-0.260 |
-0.162 |
| 156 |
247685_at |
AT5G59680
|
[Leucine-rich repeat protein kinase family protein] |
-0.260 |
0.254 |
| 157 |
256062_at |
AT1G07090
|
LSH6, LIGHT SENSITIVE HYPOCOTYLS 6 |
-0.257 |
-0.051 |
| 158 |
265272_at |
AT2G28350
|
ARF10, auxin response factor 10 |
-0.251 |
0.153 |
| 159 |
249915_at |
AT5G22870
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.247 |
-0.010 |
| 160 |
267380_at |
AT2G26170
|
CYP711A1, cytochrome P450, family 711, subfamily A, polypeptide 1, MAX1, MORE AXILLARY BRANCHES 1 |
-0.247 |
-0.185 |
| 161 |
253657_at |
AT4G30110
|
ATHMA2, ARABIDOPSIS HEAVY METAL ATPASE 2, HMA2, heavy metal atpase 2 |
-0.245 |
-0.002 |
| 162 |
263382_at |
AT2G40230
|
[HXXXD-type acyl-transferase family protein] |
-0.243 |
0.096 |
| 163 |
260170_at |
AT1G71890
|
ATSUC5, SUCROSE-PROTON SYMPORTER 5, SUC5 |
-0.241 |
-0.209 |
| 164 |
252105_at |
AT3G51470
|
[Protein phosphatase 2C family protein] |
-0.240 |
0.102 |
| 165 |
247264_at |
AT5G64530
|
ANAC104, Arabidopsis NAC domain containing protein 104, XND1, xylem NAC domain 1 |
-0.237 |
0.109 |
| 166 |
260301_at |
AT1G80290
|
[Nucleotide-diphospho-sugar transferases superfamily protein] |
-0.237 |
0.234 |
| 167 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
-0.237 |
0.007 |
| 168 |
248619_at |
AT5G49630
|
AAP6, amino acid permease 6 |
-0.236 |
-0.285 |
| 169 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.234 |
-0.396 |
| 170 |
264782_at |
AT1G08810
|
AtMYB60, myb domain protein 60, MYB60, myb domain protein 60 |
-0.234 |
-0.417 |
| 171 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.232 |
-0.119 |
| 172 |
264597_at |
AT1G04620
|
HCAR, 7-hydroxymethyl chlorophyll a (HMChl) reductase |
-0.231 |
-0.171 |
| 173 |
248484_at |
AT5G51030
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.229 |
0.050 |
| 174 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.227 |
0.050 |
| 175 |
256598_at |
AT3G30180
|
BR6OX2, brassinosteroid-6-oxidase 2, CYP85A2 |
-0.226 |
-0.878 |
| 176 |
256071_at |
AT1G13640
|
[Phosphatidylinositol 3- and 4-kinase family protein] |
-0.225 |
-0.103 |
| 177 |
261712_at |
AT1G32780
|
[GroES-like zinc-binding dehydrogenase family protein] |
-0.219 |
-0.002 |
| 178 |
253474_at |
AT4G32270
|
[Ubiquitin-like superfamily protein] |
-0.214 |
-0.078 |
| 179 |
262231_at |
AT1G68740
|
PHO1;H1 |
-0.214 |
0.092 |
| 180 |
254125_at |
AT4G24670
|
TAR2, tryptophan aminotransferase related 2 |
-0.213 |
-0.208 |
| 181 |
261388_at |
AT1G05385
|
LPA19, LOW PSII ACCUMULATION 19, Psb27-H1 |
-0.209 |
-0.182 |
| 182 |
256168_at |
AT1G51805
|
[Leucine-rich repeat protein kinase family protein] |
-0.208 |
-0.228 |
| 183 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.207 |
0.068 |
| 184 |
252972_at |
AT4G38840
|
SAUR14, SMALL AUXIN UPREGULATED RNA 14 |
-0.207 |
-0.214 |
| 185 |
262711_at |
AT1G16500
|
unknown |
-0.206 |
-0.003 |
| 186 |
260254_at |
AT1G74210
|
AtGDPD5, GDPD5, glycerophosphodiester phosphodiesterase 5 |
-0.205 |
-0.161 |
| 187 |
250485_at |
AT5G09990
|
PROPEP5, elicitor peptide 5 precursor |
-0.205 |
-0.208 |
| 188 |
257858_at |
AT3G12920
|
BRG3, BOI-related gene 3 |
-0.204 |
-0.027 |
| 189 |
249306_at |
AT5G41400
|
[RING/U-box superfamily protein] |
-0.204 |
-0.029 |
| 190 |
258003_at |
AT3G29030
|
ATEXP5, ARABIDOPSIS THALIANA EXPANSIN 5, ATEXPA5, ARABIDOPSIS THALIANA EXPANSIN A5, ATHEXP ALPHA 1.4, EXP5, EXPANSIN 5, EXPA5, expansin A5 |
-0.203 |
0.076 |
| 191 |
254068_at |
AT4G25450
|
ABCB28, ATP-binding cassette B28, ATNAP8, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN 8, NAP8, non-intrinsic ABC protein 8 |
-0.203 |
-0.249 |
| 192 |
265544_at |
AT2G28260
|
ATCNGC15, cyclic nucleotide-gated channel 15, CNGC15, cyclic nucleotide-gated channel 15 |
-0.203 |
-0.080 |
| 193 |
267063_at |
AT2G41120
|
unknown |
-0.203 |
-0.128 |
| 194 |
253421_at |
AT4G32340
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.203 |
-0.047 |
| 195 |
252619_at |
AT3G45210
|
unknown |
-0.202 |
-0.001 |
| 196 |
253296_at |
AT4G33770
|
ITPK2, inositol 1,3,4-trisphosphate 5/6 kinase 2 |
-0.201 |
-0.148 |
| 197 |
264828_at |
AT1G03380
|
ATATG18G, homolog of yeast autophagy 18 (ATG18) G, ATG18G, homolog of yeast autophagy 18 (ATG18) G |
-0.199 |
-0.020 |
| 198 |
248404_at |
AT5G51460
|
ATTPPA, TPPA, trehalose-6-phosphate phosphatase A |
-0.198 |
-0.152 |
| 199 |
265726_at |
AT2G32010
|
CVL1, CVP2 like 1 |
-0.198 |
-0.184 |
| 200 |
267590_at |
AT2G39700
|
ATEXP4, ATEXPA4, expansin A4, ATHEXP ALPHA 1.6, EXPA4, expansin A4 |
-0.198 |
-0.289 |
| 201 |
254041_at |
AT4G25830
|
[Uncharacterised protein family (UPF0497)] |
-0.198 |
-0.415 |
| 202 |
254065_at |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
-0.196 |
0.074 |
| 203 |
255578_at |
AT4G01450
|
UMAMIT30, Usually multiple acids move in and out Transporters 30 |
-0.195 |
0.015 |
| 204 |
263293_x_at |
AT2G10880
|
[hypothetical protein, similar to hypothetical protein GB:AAC26673] |
-0.194 |
0.148 |
| 205 |
251903_at |
AT3G54120
|
[Reticulon family protein] |
-0.194 |
0.092 |
| 206 |
258925_at |
AT3G10420
|
SPD1, SEEDLING PLASTID DEVELOPMENT 1 |
-0.194 |
-0.042 |
| 207 |
260167_at |
AT1G71970
|
unknown |
-0.194 |
-0.166 |
| 208 |
266325_at |
AT2G46630
|
unknown |
-0.194 |
0.055 |
| 209 |
265842_at |
AT2G35700
|
ATERF38, ERF FAMILY PROTEIN 38, ERF38, ERF family protein 38 |
-0.192 |
0.083 |
| 210 |
264843_at |
AT1G03400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.189 |
-0.030 |
| 211 |
245515_at |
AT4G15810
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.187 |
-0.132 |
| 212 |
250591_at |
AT5G07720
|
XXT3, xyloglucan xylosyltransferase 3 |
-0.187 |
0.055 |
| 213 |
261056_at |
AT1G01360
|
PYL9, PYRABACTIN RESISTANCE 1-LIKE 9, RCAR1, regulatory component of ABA receptor 1 |
-0.186 |
0.039 |
| 214 |
245417_at |
AT4G17360
|
[Formyl transferase] |
-0.185 |
-0.360 |
| 215 |
254032_at |
AT4G25940
|
[ENTH/ANTH/VHS superfamily protein] |
-0.185 |
0.116 |
| 216 |
258389_at |
AT3G15390
|
SDE5, silencing defective 5 |
-0.184 |
-0.326 |
| 217 |
262728_at |
AT1G75820
|
ATCLV1, CLV1, CLAVATA 1, FAS3, FASCIATA 3, FLO5, FLOWER DEVELOPMENT 5 |
-0.183 |
-0.322 |
| 218 |
266372_at |
AT2G41310
|
ARR8, RESPONSE REGULATOR 8, ATRR3, response regulator 3, RR3, response regulator 3 |
-0.182 |
0.212 |
| 219 |
251677_at |
AT3G56980
|
BHLH039, bHLH39, basic helix-loop-helix 39, ORG3, OBP3-RESPONSIVE GENE 3 |
-0.180 |
0.139 |
| 220 |
256819_at |
AT3G21390
|
[Mitochondrial substrate carrier family protein] |
-0.179 |
-0.098 |
| 221 |
254303_at |
AT4G22830
|
unknown |
-0.179 |
-0.049 |
| 222 |
257474_at |
AT1G80850
|
[DNA glycosylase superfamily protein] |
-0.179 |
0.063 |
| 223 |
263412_at |
AT2G28720
|
[Histone superfamily protein] |
-0.177 |
0.095 |
| 224 |
250697_at |
AT5G06800
|
[myb-like HTH transcriptional regulator family protein] |
-0.174 |
0.097 |
| 225 |
256427_at |
AT3G11090
|
LBD21, LOB domain-containing protein 21 |
-0.174 |
-0.460 |
| 226 |
261456_at |
AT1G21050
|
unknown |
-0.174 |
-0.091 |
| 227 |
262744_at |
AT1G28680
|
[HXXXD-type acyl-transferase family protein] |
-0.172 |
0.214 |
| 228 |
247701_at |
AT5G59900
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.172 |
0.058 |
| 229 |
254202_at |
AT4G24140
|
[alpha/beta-Hydrolases superfamily protein] |
-0.172 |
0.086 |
| 230 |
261151_at |
AT1G19650
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.171 |
0.000 |
| 231 |
266882_at |
AT2G44670
|
unknown |
-0.171 |
-0.012 |
| 232 |
248097_at |
AT5G55260
|
PPX-2, PROTEIN PHOSPHATASE X -2, PPX2, protein phosphatase X 2 |
-0.171 |
-0.034 |
| 233 |
247831_at |
AT5G58540
|
[Protein kinase superfamily protein] |
-0.171 |
0.513 |
| 234 |
265722_at |
AT2G40100
|
LHCB4.3, light harvesting complex photosystem II |
-0.171 |
-0.251 |
| 235 |
247691_at |
AT5G59720
|
HSP18.2, heat shock protein 18.2 |
-0.170 |
0.225 |
| 236 |
264841_at |
AT1G03740
|
[Protein kinase superfamily protein] |
-0.170 |
0.140 |
| 237 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.170 |
-0.254 |