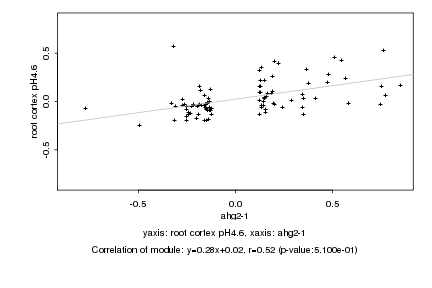
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ahg2-1 |
Log2 signal ratio
root cortex pH4.6 |
| 1 |
244901_at |
ATMG00640
|
ORF25 |
0.848 |
0.173 |
| 2 |
244943_at |
ATMG00070
|
NAD9, NADH dehydrogenase subunit 9 |
0.772 |
0.065 |
| 3 |
250515_at |
AT5G09570
|
[Cox19-like CHCH family protein] |
0.761 |
0.533 |
| 4 |
244945_at |
ATMG00110
|
ABCI2, ATP-binding cassette I2, CCB206, cytochrome C biogenesis 206 |
0.752 |
0.164 |
| 5 |
257333_at |
ATMG01360
|
COX1, cytochrome oxidase |
0.743 |
-0.021 |
| 6 |
244925_at |
ATMG00510
|
NAD7, NADH dehydrogenase subunit 7 |
0.582 |
-0.019 |
| 7 |
254419_at |
AT4G21490
|
NDB3, NAD(P)H dehydrogenase B3 |
0.564 |
0.239 |
| 8 |
244902_at |
ATMG00650
|
NAD4L, NADH dehydrogenase subunit 4L |
0.543 |
0.428 |
| 9 |
263515_at |
AT2G21640
|
unknown |
0.507 |
0.465 |
| 10 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.479 |
0.281 |
| 11 |
265422_at |
AT2G20800
|
NDB4, NAD(P)H dehydrogenase B4 |
0.469 |
0.203 |
| 12 |
254286_at |
AT4G22950
|
AGL19, AGAMOUS-like 19, GL19 |
0.408 |
0.036 |
| 13 |
244921_s_at |
ATMG01000
|
ORF114 |
0.374 |
0.196 |
| 14 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.363 |
0.336 |
| 15 |
246888_at |
AT5G26270
|
unknown |
0.350 |
-0.131 |
| 16 |
256459_at |
AT1G36180
|
ACC2, acetyl-CoA carboxylase 2 |
0.346 |
0.033 |
| 17 |
266729_at |
AT2G03130
|
[Ribosomal protein L12/ ATP-dependent Clp protease adaptor protein ClpS family protein] |
0.344 |
0.077 |
| 18 |
249784_at |
AT5G24280
|
GMI1, GAMMA-IRRADIATION AND MITOMYCIN C INDUCED 1 |
0.343 |
-0.055 |
| 19 |
256188_at |
AT1G30160
|
unknown |
0.287 |
0.015 |
| 20 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.240 |
-0.061 |
| 21 |
264868_at |
AT1G24095
|
[Putative thiol-disulphide oxidoreductase DCC] |
0.219 |
0.397 |
| 22 |
254415_at |
AT4G21326
|
ATSBT3.12, subtilase 3.12, SBT3.12, subtilase 3.12 |
0.198 |
-0.022 |
| 23 |
262459_at |
AT1G50400
|
[Eukaryotic porin family protein] |
0.197 |
0.423 |
| 24 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
0.193 |
-0.016 |
| 25 |
252383_at |
AT3G47780
|
ABCA7, ATP-binding cassette A7, ATATH6, A. THALIANA ABC2 HOMOLOG 6, ATH6, ABC2 homolog 6 |
0.187 |
0.105 |
| 26 |
252864_at |
AT4G39740
|
HCC2, homologue of copper chaperone SCO1 2 |
0.186 |
0.264 |
| 27 |
264830_at |
AT1G03710
|
[Cystatin/monellin superfamily protein] |
0.183 |
0.092 |
| 28 |
251871_at |
AT3G54520
|
unknown |
0.162 |
0.090 |
| 29 |
257336_at |
ATMG01410
|
ORF204, open reading frame 204 |
0.159 |
0.062 |
| 30 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
0.150 |
-0.112 |
| 31 |
266738_at |
AT2G47010
|
unknown |
0.150 |
-0.082 |
| 32 |
247339_at |
AT5G63690
|
[Nucleic acid-binding, OB-fold-like protein] |
0.149 |
0.033 |
| 33 |
248101_at |
AT5G55200
|
MGE1, mitochondrial GrpE 1 |
0.146 |
0.225 |
| 34 |
247167_at |
AT5G65850
|
[F-box and associated interaction domains-containing protein] |
0.146 |
0.048 |
| 35 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
0.142 |
0.004 |
| 36 |
249384_at |
AT5G39890
|
unknown |
0.140 |
-0.038 |
| 37 |
245879_at |
AT5G09420
|
AtmtOM64, ATTOC64-V, translocon at the outer membrane of chloroplasts 64-V, MTOM64, ARABIDOPSIS THALIANA TRANSLOCON AT THE OUTER MEMBRANE OF CHLOROPLASTS 64-V, OM64, outer membrane 64, TOC64-V, translocon at the outer membrane of chloroplasts 64-V |
0.134 |
0.358 |
| 38 |
257474_at |
AT1G80850
|
[DNA glycosylase superfamily protein] |
0.132 |
-0.052 |
| 39 |
260933_at |
AT1G02470
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.131 |
-0.041 |
| 40 |
258009_at |
AT3G19440
|
[Pseudouridine synthase family protein] |
0.129 |
0.158 |
| 41 |
249258_at |
AT5G41650
|
[Lactoylglutathione lyase / glyoxalase I family protein] |
0.129 |
0.096 |
| 42 |
245951_at |
AT5G19550
|
AAT2, ASPARTATE AMINOTRANSFERASE 2, ASP2, aspartate aminotransferase 2 |
0.126 |
0.220 |
| 43 |
245893_at |
AT5G09270
|
unknown |
0.122 |
0.159 |
| 44 |
258380_at |
AT3G16650
|
PRL2, Pleiotropic regulatory locus 2 |
0.121 |
0.095 |
| 45 |
265926_at |
AT2G18600
|
[Ubiquitin-conjugating enzyme family protein] |
0.121 |
0.011 |
| 46 |
246249_at |
AT4G36680
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.119 |
0.327 |
| 47 |
247616_at |
AT5G60260
|
unknown |
0.119 |
-0.124 |
| 48 |
253301_at |
AT4G33720
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.776 |
-0.062 |
| 49 |
255516_at |
AT4G02270
|
RHS13, root hair specific 13 |
-0.495 |
-0.246 |
| 50 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
-0.333 |
-0.011 |
| 51 |
257481_at |
AT1G08430
|
ALMT1, aluminum-activated malate transporter 1, ATALMT1, ARABIDOPSIS THALIANA ALUMINUM-ACTIVATED MALATE TRANSPORTER 1 |
-0.319 |
0.578 |
| 52 |
255814_at |
AT1G19900
|
[glyoxal oxidase-related protein] |
-0.314 |
-0.191 |
| 53 |
251448_at |
AT3G59845
|
[Zinc-binding dehydrogenase family protein] |
-0.310 |
-0.050 |
| 54 |
262760_at |
AT1G10770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.275 |
0.021 |
| 55 |
262004_at |
AT1G64480
|
CBL8, calcineurin B-like protein 8 |
-0.273 |
-0.041 |
| 56 |
260668_at |
AT1G19530
|
unknown |
-0.267 |
-0.025 |
| 57 |
256352_at |
AT1G54970
|
ATPRP1, proline-rich protein 1, PRP1, proline-rich protein 1, RHS7, ROOT HAIR SPECIFIC 7 |
-0.257 |
-0.149 |
| 58 |
264138_at |
AT1G78950
|
AtBAS, AtLUP4, BAS, beta-amyrin synthase |
-0.256 |
-0.049 |
| 59 |
253244_at |
AT4G34580
|
AtSFH1, COW1, CAN OF WORMS1, SRH1, SHORT ROOT HAIR 1 |
-0.255 |
-0.074 |
| 60 |
259525_at |
AT1G12560
|
ATEXP7, ATEXPA7, expansin A7, ATHEXP ALPHA 1.26, EXP7, EXPA7, expansin A7 |
-0.253 |
-0.196 |
| 61 |
248252_at |
AT5G53250
|
AGP22, arabinogalactan protein 22, ATAGP22, ARABINOGALACTAN PROTEIN 22 |
-0.245 |
-0.131 |
| 62 |
265111_at |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.244 |
-0.111 |
| 63 |
253774_at |
AT4G28530
|
anac074, NAC domain containing protein 74, NAC074, NAC domain containing protein 74 |
-0.233 |
-0.120 |
| 64 |
264734_at |
AT1G62280
|
SLAH1, SLAC1 homologue 1 |
-0.229 |
-0.050 |
| 65 |
248926_at |
AT5G45880
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.220 |
-0.029 |
| 66 |
256584_at |
AT3G28750
|
unknown |
-0.220 |
-0.041 |
| 67 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
-0.203 |
-0.167 |
| 68 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.199 |
-0.042 |
| 69 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
-0.194 |
-0.035 |
| 70 |
252447_at |
AT3G47040
|
[Glycosyl hydrolase family protein] |
-0.192 |
-0.126 |
| 71 |
267254_at |
AT2G23030
|
SNRK2-9, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-9, SNRK2.9, SNF1-related protein kinase 2.9 |
-0.187 |
-0.026 |
| 72 |
255891_at |
AT1G17870
|
ATEGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3, EGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3 |
-0.186 |
0.163 |
| 73 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
-0.185 |
0.114 |
| 74 |
246425_at |
AT5G17420
|
ATCESA7, CESA7, CELLULOSE SYNTHASE CATALYTIC SUBUNIT 7, IRX3, IRREGULAR XYLEM 3, MUR10, MURUS 10 |
-0.175 |
-0.035 |
| 75 |
261991_at |
AT1G33700
|
[Beta-glucosidase, GBA2 type family protein] |
-0.164 |
0.069 |
| 76 |
264590_at |
AT2G17710
|
unknown |
-0.164 |
-0.196 |
| 77 |
248856_at |
AT5G46620
|
unknown |
-0.164 |
-0.033 |
| 78 |
263614_at |
AT2G25240
|
AtCCP3, CCP3, conserved in ciliated species and in the land plants 3 |
-0.157 |
-0.029 |
| 79 |
256283_at |
AT3G12540
|
unknown |
-0.156 |
-0.060 |
| 80 |
266344_at |
AT2G01580
|
unknown |
-0.154 |
-0.078 |
| 81 |
266690_at |
AT2G19900
|
ATNADP-ME1, Arabidopsis thaliana NADP-malic enzyme 1, NADP-ME1, NADP-malic enzyme 1 |
-0.154 |
-0.072 |
| 82 |
258367_at |
AT3G14370
|
WAG2 |
-0.153 |
-0.190 |
| 83 |
246868_at |
AT5G26010
|
[Protein phosphatase 2C family protein] |
-0.149 |
-0.008 |
| 84 |
256181_at |
AT1G51820
|
[Leucine-rich repeat protein kinase family protein] |
-0.148 |
-0.016 |
| 85 |
246251_at |
AT4G37220
|
[Cold acclimation protein WCOR413 family] |
-0.145 |
-0.091 |
| 86 |
247765_at |
AT5G58860
|
CYP86, CYP86A1, cytochrome P450, family 86, subfamily A, polypeptide 1, HORST, hydroxylase of root suberized tissue |
-0.143 |
0.040 |
| 87 |
254263_at |
AT4G23493
|
unknown |
-0.139 |
-0.177 |
| 88 |
256799_at |
AT3G18560
|
unknown |
-0.136 |
-0.052 |
| 89 |
250277_at |
AT5G12940
|
[Leucine-rich repeat (LRR) family protein] |
-0.136 |
-0.087 |
| 90 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
-0.134 |
0.006 |
| 91 |
262743_at |
AT1G29020
|
[Calcium-binding EF-hand family protein] |
-0.132 |
-0.083 |
| 92 |
246943_at |
AT5G25440
|
[Protein kinase superfamily protein] |
-0.131 |
0.129 |
| 93 |
252834_at |
AT4G40070
|
[RING/U-box superfamily protein] |
-0.128 |
-0.067 |
| 94 |
248916_at |
AT5G45840
|
[Leucine-rich repeat protein kinase family protein] |
-0.126 |
-0.133 |