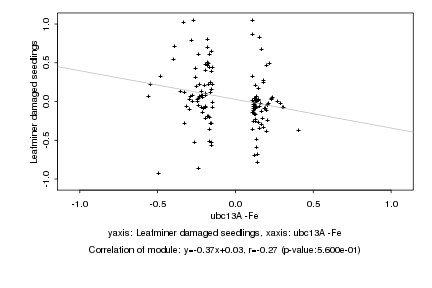
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ubc13A -Fe |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
0.400 |
-0.371 |
| 2 |
257823_at |
AT3G25190
|
[Vacuolar iron transporter (VIT) family protein] |
0.308 |
-0.074 |
| 3 |
245007_at |
ATCG00350
|
PSAA |
0.296 |
-0.070 |
| 4 |
254888_at |
AT4G11780
|
TRM10, TON1 Recruiting Motif 10 |
0.287 |
-0.025 |
| 5 |
247477_at |
AT5G62340
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.267 |
0.006 |
| 6 |
260213_at |
AT1G74490
|
[Protein kinase superfamily protein] |
0.238 |
0.053 |
| 7 |
260813_at |
AT1G43700
|
SUE3, sulphate utilization efficiency 3, VIP1, VIRE2-interacting protein 1 |
0.229 |
0.036 |
| 8 |
260067_at |
AT1G73780
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.226 |
0.040 |
| 9 |
266072_at |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
0.217 |
0.496 |
| 10 |
245006_at |
ATCG00340
|
PSAB |
0.207 |
-0.022 |
| 11 |
244937_at |
ATCG01110
|
NDHH, NAD(P)H dehydrogenase subunit H |
0.203 |
-0.243 |
| 12 |
244961_at |
ATCG01040
|
YCF5 |
0.200 |
-0.045 |
| 13 |
266141_at |
AT2G02120
|
LCR70, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 70, PDF2.1 |
0.200 |
-0.031 |
| 14 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
0.197 |
0.473 |
| 15 |
258367_at |
AT3G14370
|
WAG2 |
0.197 |
-0.107 |
| 16 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
0.194 |
-0.381 |
| 17 |
264598_at |
AT1G04610
|
YUC3, YUCCA 3 |
0.192 |
-0.086 |
| 18 |
245010_at |
ATCG00420
|
NDHJ, NADH dehydrogenase subunit J |
0.185 |
-0.085 |
| 19 |
262236_at |
AT1G48330
|
unknown |
0.178 |
0.278 |
| 20 |
262277_at |
AT1G68650
|
[Uncharacterized protein family (UPF0016)] |
0.175 |
0.258 |
| 21 |
254878_at |
AT4G11660
|
AT-HSFB2B, HSF7, HSFB2B, HEAT SHOCK TRANSCRIPTION FACTOR B2B |
0.175 |
-0.323 |
| 22 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
0.173 |
-0.212 |
| 23 |
252692_at |
AT3G43960
|
[Cysteine proteinases superfamily protein] |
0.167 |
-0.293 |
| 24 |
248040_at |
AT5G55970
|
[RING/U-box superfamily protein] |
0.165 |
-0.125 |
| 25 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
0.163 |
0.675 |
| 26 |
252501_at |
AT3G46880
|
unknown |
0.160 |
-0.024 |
| 27 |
246591_at |
AT5G14880
|
KUP8, potassium uptake 8 |
0.153 |
-0.342 |
| 28 |
262756_at |
AT1G16370
|
ATOCT6, ARABIDOPSIS THALIANA ORGANIC CATION/CARNITINE TRANSPORTER 6, OCT6, organic cation/carnitine transporter 6 |
0.153 |
0.830 |
| 29 |
260558_at |
AT2G43600
|
[Chitinase family protein] |
0.153 |
-0.053 |
| 30 |
252047_at |
AT3G52490
|
SMXL3, SMAX1-like 3 |
0.147 |
-0.264 |
| 31 |
250802_at |
AT5G04970
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.146 |
0.022 |
| 32 |
266710_at |
AT2G46850
|
[Protein kinase superfamily protein] |
0.145 |
0.032 |
| 33 |
263610_at |
AT2G16230
|
[O-Glycosyl hydrolases family 17 protein] |
0.144 |
0.031 |
| 34 |
261026_at |
AT1G01240
|
unknown |
0.143 |
0.172 |
| 35 |
262730_at |
AT1G16390
|
ATOCT3, organic cation/carnitine transporter 3, OCT3, organic cation/carnitine transporter 3 |
0.139 |
-0.678 |
| 36 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
0.136 |
-0.780 |
| 37 |
255129_at |
AT4G08290
|
UMAMIT20, Usually multiple acids move in and out Transporters 20 |
0.135 |
0.038 |
| 38 |
244999_at |
ATCG00190
|
RPOB, RNA polymerase subunit beta |
0.135 |
-0.075 |
| 39 |
246703_at |
AT5G28080
|
WNK9 |
0.132 |
0.032 |
| 40 |
259264_at |
AT3G01260
|
[Galactose mutarotase-like superfamily protein] |
0.132 |
-0.076 |
| 41 |
255028_at |
AT4G09890
|
unknown |
0.131 |
-0.490 |
| 42 |
249305_at |
AT5G41490
|
[F-box associated ubiquitination effector family protein] |
0.130 |
0.003 |
| 43 |
257203_at |
AT3G23730
|
XTH16, xyloglucan endotransglucosylase/hydrolase 16 |
0.129 |
-0.589 |
| 44 |
253155_at |
AT4G35720
|
unknown |
0.129 |
0.075 |
| 45 |
264138_at |
AT1G78950
|
AtBAS, AtLUP4, BAS, beta-amyrin synthase |
0.128 |
-0.012 |
| 46 |
266019_at |
AT2G18750
|
[Calmodulin-binding protein] |
0.128 |
0.210 |
| 47 |
250803_at |
AT5G04980
|
[DNAse I-like superfamily protein] |
0.128 |
0.041 |
| 48 |
251617_at |
AT3G58000
|
[VQ motif-containing protein] |
0.127 |
-0.085 |
| 49 |
251900_at |
AT3G54430
|
SRS6, SHI-related sequence 6 |
0.125 |
-0.257 |
| 50 |
245674_at |
AT1G56680
|
[Chitinase family protein] |
0.124 |
0.021 |
| 51 |
266172_at |
AT2G39010
|
PIP2;6, PLASMA MEMBRANE INTRINSIC PROTEIN 2;6, PIP2E, plasma membrane intrinsic protein 2E |
0.124 |
-0.060 |
| 52 |
257745_at |
AT3G29240
|
unknown |
0.124 |
-0.167 |
| 53 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
0.123 |
-0.227 |
| 54 |
257181_at |
AT3G13190
|
unknown |
0.122 |
-0.147 |
| 55 |
255305_at |
AT4G04770
|
ABC1, ATP binding cassette protein 1, ABCI8, ATP-binding cassette I8, ATABC1, ATP binding cassette protein 1, ATNAP1, ARABIDOPSIS THALIANA NUCLEOSOME ASSEMBLY PROTEIN 1, LAF6, LONG AFTER FR |
0.120 |
-0.079 |
| 56 |
247457_at |
AT5G62170
|
TRM25, TON1 Recruiting Motif 25 |
0.118 |
-0.155 |
| 57 |
263164_at |
AT1G03070
|
AtLFG4, LFG4, LIFEGUARD 4 |
0.118 |
-0.035 |
| 58 |
249336_at |
AT5G41070
|
DRB5, dsRNA-binding protein 5 |
0.117 |
0.043 |
| 59 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
0.117 |
-0.688 |
| 60 |
254206_at |
AT4G24180
|
ATTLP1, TLP1, THAUMATIN-LIKE PROTEIN 1 |
0.117 |
-0.059 |
| 61 |
249978_at |
AT5G18850
|
unknown |
0.113 |
-0.038 |
| 62 |
250145_at |
AT5G14690
|
unknown |
0.113 |
-0.008 |
| 63 |
246765_at |
AT5G27330
|
[Prefoldin chaperone subunit family protein] |
0.113 |
-0.085 |
| 64 |
244986_at |
ATCG00820
|
RPS19, ribosomal protein S19 |
0.112 |
-0.064 |
| 65 |
253786_at |
AT4G28650
|
[Leucine-rich repeat transmembrane protein kinase family protein] |
0.112 |
-0.105 |
| 66 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
0.111 |
-0.256 |
| 67 |
254387_at |
AT4G21850
|
ATMSRB9, methionine sulfoxide reductase B9, MSRB9, methionine sulfoxide reductase B9 |
0.109 |
0.868 |
| 68 |
246229_at |
AT4G37160
|
sks15, SKU5 similar 15 |
0.109 |
0.018 |
| 69 |
252175_at |
AT3G50700
|
AtIDD2, indeterminate(ID)-domain 2, IDD2, indeterminate(ID)-domain 2 |
0.109 |
0.332 |
| 70 |
261394_at |
AT1G79680
|
ATWAKL10, WAKL10, WALL ASSOCIATED KINASE (WAK)-LIKE 10 |
0.109 |
1.058 |
| 71 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
0.109 |
-0.358 |
| 72 |
260855_at |
AT1G21920
|
[Histone H3 K4-specific methyltransferase SET7/9 family protein] |
0.108 |
-0.140 |
| 73 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
-0.562 |
0.073 |
| 74 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.549 |
0.224 |
| 75 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-0.499 |
-0.918 |
| 76 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.484 |
0.324 |
| 77 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
-0.402 |
0.545 |
| 78 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
-0.397 |
0.717 |
| 79 |
255883_at |
AT1G20270
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.358 |
0.131 |
| 80 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
-0.339 |
1.022 |
| 81 |
247463_at |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
-0.331 |
-0.277 |
| 82 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
-0.331 |
0.120 |
| 83 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.318 |
-0.057 |
| 84 |
245560_at |
AT4G15480
|
UGT84A1 |
-0.299 |
0.032 |
| 85 |
250558_at |
AT5G07990
|
CYP75B1, CYTOCHROME P450 75B1, D501, TT7, TRANSPARENT TESTA 7 |
-0.296 |
-0.100 |
| 86 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
-0.293 |
0.065 |
| 87 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
-0.286 |
0.795 |
| 88 |
252517_at |
AT3G46340
|
[Leucine-rich repeat protein kinase family protein] |
-0.281 |
0.002 |
| 89 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
-0.276 |
0.082 |
| 90 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.270 |
1.055 |
| 91 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
-0.268 |
-0.526 |
| 92 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
-0.263 |
0.437 |
| 93 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
-0.260 |
0.320 |
| 94 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.251 |
0.201 |
| 95 |
265422_at |
AT2G20800
|
NDB4, NAD(P)H dehydrogenase B4 |
-0.249 |
0.006 |
| 96 |
262092_at |
AT1G56150
|
SAUR71, SMALL AUXIN UPREGULATED 71 |
-0.244 |
0.030 |
| 97 |
261907_at |
AT1G65060
|
4CL3, 4-coumarate:CoA ligase 3 |
-0.243 |
-0.864 |
| 98 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
-0.242 |
-0.040 |
| 99 |
262629_at |
AT1G06460
|
ACD31.2, ALPHA-CRYSTALLIN DOMAIN 31.2, ACD32.1, alpha-crystallin domain 32.1 |
-0.241 |
0.047 |
| 100 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
-0.240 |
0.609 |
| 101 |
248625_at |
AT5G48880
|
KAT5, 3-KETO-ACYL-COENZYME A THIOLASE 5, PKT1, PEROXISOMAL-3-KETO-ACYL-COA THIOLASE 1, PKT2, peroxisomal 3-keto-acyl-CoA thiolase 2 |
-0.232 |
0.221 |
| 102 |
259788_at |
AT1G29670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.231 |
0.075 |
| 103 |
263515_at |
AT2G21640
|
unknown |
-0.223 |
0.136 |
| 104 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
-0.220 |
0.086 |
| 105 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.219 |
-0.073 |
| 106 |
260522_x_at |
AT2G41730
|
unknown |
-0.218 |
0.082 |
| 107 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.215 |
0.063 |
| 108 |
263761_at |
AT2G21330
|
AtFBA1, FBA1, fructose-bisphosphate aldolase 1 |
-0.212 |
-0.135 |
| 109 |
266738_at |
AT2G47010
|
unknown |
-0.208 |
-0.079 |
| 110 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
-0.204 |
0.213 |
| 111 |
250702_at |
AT5G06730
|
[Peroxidase superfamily protein] |
-0.198 |
0.111 |
| 112 |
264635_at |
AT1G65500
|
unknown |
-0.198 |
0.405 |
| 113 |
254998_at |
AT4G09760
|
[Protein kinase superfamily protein] |
-0.197 |
0.086 |
| 114 |
256503_at |
AT1G75250
|
ATRL6, RAD-like 6, RL6, RAD-like 6, RSM3, RADIALIS-LIKE SANT/MYB 3 |
-0.197 |
-0.065 |
| 115 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
-0.196 |
0.478 |
| 116 |
251827_at |
AT3G55120
|
A11, AtCHI, CFI, CHALCONE FLAVANONE ISOMERASE, CHI, chalcone isomerase, TT5, TRANSPARENT TESTA 5 |
-0.194 |
-0.213 |
| 117 |
254460_at |
AT4G21210
|
ATRP1, PPDK regulatory protein, RP1, PPDK regulatory protein |
-0.193 |
-0.064 |
| 118 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
-0.185 |
0.701 |
| 119 |
249750_at |
AT5G24570
|
unknown |
-0.184 |
-0.191 |
| 120 |
260051_at |
AT1G78210
|
[alpha/beta-Hydrolases superfamily protein] |
-0.183 |
0.472 |
| 121 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
-0.183 |
0.509 |
| 122 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
-0.181 |
0.806 |
| 123 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
-0.179 |
-0.187 |
| 124 |
250646_at |
AT5G06720
|
ATPA2, peroxidase 2, AtPRX53, PA2, peroxidase 2, PRX53, peroxidase 53 |
-0.178 |
0.230 |
| 125 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
-0.175 |
0.501 |
| 126 |
249767_at |
AT5G24090
|
ATCHIA, chitinase A, CHIA, chitinase A |
-0.171 |
0.442 |
| 127 |
261921_at |
AT1G65900
|
unknown |
-0.169 |
-0.205 |
| 128 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-0.169 |
-0.503 |
| 129 |
250149_at |
AT5G14700
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.168 |
0.615 |
| 130 |
254020_at |
AT4G25700
|
B1, BCH1, BETA CAROTENOID HYDROXYLASE 1, BETA-OHASE 1, beta-hydroxylase 1, chy1 |
-0.167 |
-0.359 |
| 131 |
266108_at |
AT2G37900
|
[Major facilitator superfamily protein] |
-0.165 |
0.255 |
| 132 |
256714_at |
AT2G34080
|
[Cysteine proteinases superfamily protein] |
-0.161 |
0.125 |
| 133 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
-0.161 |
-0.281 |
| 134 |
256489_at |
AT1G31550
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.160 |
0.395 |
| 135 |
253485_at |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
-0.160 |
0.656 |
| 136 |
255016_at |
AT4G10120
|
ATSPS4F |
-0.159 |
-0.557 |
| 137 |
254024_at |
AT4G25780
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.158 |
-0.279 |
| 138 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
-0.157 |
-0.524 |
| 139 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
-0.154 |
0.156 |
| 140 |
261125_at |
AT1G04990
|
[Zinc finger C-x8-C-x5-C-x3-H type family protein] |
-0.152 |
0.226 |
| 141 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
-0.151 |
0.446 |
| 142 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.151 |
-0.075 |
| 143 |
254404_at |
AT4G21340
|
B70 |
-0.150 |
-0.008 |