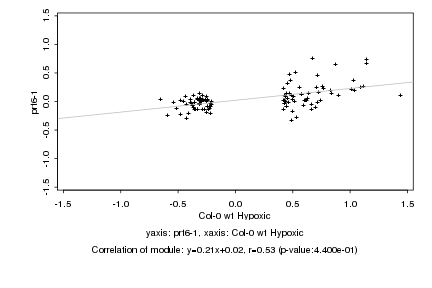
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Col-0 wt Hypoxic |
Log2 signal ratio
prt6-1 |
| 1 |
250464_at |
AT5G10040
|
unknown |
1.436 |
0.121 |
| 2 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
1.141 |
0.668 |
| 3 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
1.135 |
0.746 |
| 4 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
1.109 |
0.265 |
| 5 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
1.084 |
0.257 |
| 6 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
1.036 |
0.206 |
| 7 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
1.026 |
0.383 |
| 8 |
254200_at |
AT4G24110
|
unknown |
1.011 |
0.216 |
| 9 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
0.898 |
0.120 |
| 10 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
0.872 |
0.658 |
| 11 |
257153_at |
AT3G27220
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.831 |
0.144 |
| 12 |
257925_at |
AT3G23170
|
unknown |
0.825 |
0.202 |
| 13 |
247024_at |
AT5G66985
|
unknown |
0.765 |
0.235 |
| 14 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.753 |
0.279 |
| 15 |
253573_at |
AT4G31020
|
[alpha/beta-Hydrolases superfamily protein] |
0.737 |
0.022 |
| 16 |
249384_at |
AT5G39890
|
unknown |
0.724 |
0.167 |
| 17 |
253060_at |
AT4G37710
|
[VQ motif-containing protein] |
0.714 |
-0.003 |
| 18 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
0.711 |
0.456 |
| 19 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
0.704 |
0.258 |
| 20 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
0.691 |
-0.098 |
| 21 |
245226_at |
AT3G29970
|
[B12D protein] |
0.670 |
0.759 |
| 22 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
0.658 |
-0.047 |
| 23 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
0.656 |
-0.124 |
| 24 |
245401_at |
AT4G17670
|
unknown |
0.634 |
0.150 |
| 25 |
266486_at |
AT2G47950
|
unknown |
0.625 |
0.061 |
| 26 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.615 |
0.019 |
| 27 |
263295_at |
AT2G14210
|
AGL44, AGAMOUS-like 44, ANR1, ARABIDOPSIS NITRATE REGULATED 1 |
0.607 |
0.032 |
| 28 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
0.597 |
0.033 |
| 29 |
260451_at |
AT1G72360
|
AtERF73, ERF73, ethylene response factor 73, HRE1, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 1 |
0.588 |
-0.056 |
| 30 |
248138_at |
AT5G54960
|
PDC2, pyruvate decarboxylase-2 |
0.574 |
0.123 |
| 31 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
0.572 |
0.135 |
| 32 |
245586_at |
AT4G14980
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.555 |
0.253 |
| 33 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
0.531 |
-0.264 |
| 34 |
245998_at |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
0.522 |
0.509 |
| 35 |
262124_at |
AT1G59660
|
[Nucleoporin autopeptidase] |
0.508 |
0.011 |
| 36 |
255900_at |
AT1G17830
|
unknown |
0.499 |
0.102 |
| 37 |
249015_at |
AT5G44730
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.492 |
0.036 |
| 38 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
0.489 |
-0.173 |
| 39 |
250472_at |
AT5G10210
|
unknown |
0.488 |
0.119 |
| 40 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.483 |
-0.322 |
| 41 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.472 |
0.373 |
| 42 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
0.470 |
0.481 |
| 43 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
0.464 |
0.155 |
| 44 |
259033_at |
AT3G09405
|
[Pectinacetylesterase family protein] |
0.460 |
-0.015 |
| 45 |
261567_at |
AT1G33055
|
unknown |
0.450 |
0.325 |
| 46 |
257922_at |
AT3G23150
|
ETR2, ethylene response 2 |
0.450 |
0.053 |
| 47 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.444 |
-0.071 |
| 48 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
0.444 |
-0.079 |
| 49 |
248719_at |
AT5G47910
|
ATRBOHD, RBOHD, respiratory burst oxidase homologue D |
0.437 |
0.144 |
| 50 |
266965_at |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
0.432 |
0.010 |
| 51 |
267276_at |
AT2G30130
|
ASL5, LBD12, PCK1, PEACOCK 1 |
0.428 |
0.095 |
| 52 |
253987_at |
AT4G26270
|
PFK3, phosphofructokinase 3 |
0.427 |
0.029 |
| 53 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.425 |
-0.026 |
| 54 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
0.425 |
-0.029 |
| 55 |
259616_at |
AT1G47960
|
ATC/VIF1, CELL WALL / VACUOLAR INHIBITOR OF FRUCTOSIDASE 1, C/VIF1, cell wall / vacuolar inhibitor of fructosidase 1 |
0.421 |
0.121 |
| 56 |
253404_at |
AT4G32840
|
PFK6, phosphofructokinase 6 |
0.417 |
0.034 |
| 57 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.416 |
-0.127 |
| 58 |
258879_at |
AT3G03270
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.414 |
0.229 |
| 59 |
259264_at |
AT3G01260
|
[Galactose mutarotase-like superfamily protein] |
-0.658 |
0.048 |
| 60 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
-0.595 |
-0.231 |
| 61 |
256352_at |
AT1G54970
|
ATPRP1, proline-rich protein 1, PRP1, proline-rich protein 1, RHS7, ROOT HAIR SPECIFIC 7 |
-0.548 |
-0.008 |
| 62 |
254056_at |
AT4G25250
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.521 |
-0.106 |
| 63 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
-0.483 |
-0.227 |
| 64 |
252638_at |
AT3G44540
|
FAR4, fatty acid reductase 4 |
-0.483 |
0.021 |
| 65 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
-0.456 |
0.006 |
| 66 |
261956_at |
AT1G64590
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.438 |
0.096 |
| 67 |
250541_at |
AT5G09520
|
PELPK2, Pro-Glu-Leu|Ile|Val-Pro-Lys 2 |
-0.432 |
-0.039 |
| 68 |
247878_at |
AT5G57760
|
unknown |
-0.431 |
-0.281 |
| 69 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
-0.416 |
-0.198 |
| 70 |
265846_at |
AT2G35770
|
scpl28, serine carboxypeptidase-like 28 |
-0.395 |
0.039 |
| 71 |
250801_at |
AT5G04960
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.394 |
-0.008 |
| 72 |
253155_at |
AT4G35720
|
unknown |
-0.389 |
-0.024 |
| 73 |
246229_at |
AT4G37160
|
sks15, SKU5 similar 15 |
-0.378 |
-0.061 |
| 74 |
264318_at |
AT1G04220
|
KCS2, 3-ketoacyl-CoA synthase 2 |
-0.378 |
-0.054 |
| 75 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
-0.368 |
-0.096 |
| 76 |
258037_at |
AT3G21230
|
4CL5, 4-coumarate:CoA ligase 5 |
-0.366 |
0.115 |
| 77 |
253353_at |
AT4G33730
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.363 |
-0.001 |
| 78 |
253309_at |
AT4G33790
|
CER4, ECERIFERUM 4, FAR3, FATTY ACID REDUCTASE 3, G7 |
-0.361 |
-0.010 |
| 79 |
265102_at |
AT1G30870
|
[Peroxidase superfamily protein] |
-0.359 |
-0.136 |
| 80 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
-0.356 |
-0.135 |
| 81 |
259525_at |
AT1G12560
|
ATEXP7, ATEXPA7, expansin A7, ATHEXP ALPHA 1.26, EXP7, EXPA7, expansin A7 |
-0.337 |
0.053 |
| 82 |
264010_at |
AT2G21100
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.336 |
0.039 |
| 83 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
-0.334 |
-0.132 |
| 84 |
255808_at |
AT4G10280
|
[RmlC-like cupins superfamily protein] |
-0.333 |
-0.130 |
| 85 |
249289_at |
AT5G41040
|
ASFT, aliphatic suberin feruloyl-transferase, HHT, hydroxycinnamoyl- CoA:ω-hydroxyacid O-hydroxycinnamoyltransferase, RWP1, REDUCED LEVELS OF WALL-BOUND PHENOLICS 1 |
-0.321 |
-0.020 |
| 86 |
254836_at |
AT4G12330
|
CYP706A7, cytochrome P450, family 706, subfamily A, polypeptide 7 |
-0.320 |
-0.038 |
| 87 |
266191_at |
AT2G39040
|
[Peroxidase superfamily protein] |
-0.320 |
-0.042 |
| 88 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
-0.319 |
0.142 |
| 89 |
257105_at |
AT3G15300
|
[VQ motif-containing protein] |
-0.317 |
-0.022 |
| 90 |
253301_at |
AT4G33720
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.316 |
0.026 |
| 91 |
262198_at |
AT1G53830
|
ATPME2, pectin methylesterase 2, PME2, pectin methylesterase 2 |
-0.315 |
0.064 |
| 92 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
-0.312 |
0.006 |
| 93 |
248895_at |
AT5G46330
|
FLS2, FLAGELLIN-SENSITIVE 2 |
-0.312 |
0.057 |
| 94 |
250778_at |
AT5G05500
|
MOP10 |
-0.300 |
0.012 |
| 95 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.292 |
-0.123 |
| 96 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
-0.292 |
0.050 |
| 97 |
266828_at |
AT2G22930
|
[UDP-Glycosyltransferase superfamily protein] |
-0.288 |
0.116 |
| 98 |
267372_at |
AT2G26290
|
ARSK1, root-specific kinase 1 |
-0.287 |
0.026 |
| 99 |
248118_at |
AT5G55050
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.271 |
-0.126 |
| 100 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
-0.266 |
0.036 |
| 101 |
246203_at |
AT4G36610
|
[alpha/beta-Hydrolases superfamily protein] |
-0.255 |
0.012 |
| 102 |
253734_at |
AT4G29180
|
RHS16, root hair specific 16 |
-0.254 |
0.098 |
| 103 |
253667_at |
AT4G30170
|
[Peroxidase family protein] |
-0.253 |
-0.189 |
| 104 |
251254_at |
AT3G62270
|
BOR2, REQUIRES HIGH BORON 2 |
-0.252 |
-0.085 |
| 105 |
248252_at |
AT5G53250
|
AGP22, arabinogalactan protein 22, ATAGP22, ARABINOGALACTAN PROTEIN 22 |
-0.251 |
-0.077 |
| 106 |
254326_at |
AT4G22610
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.251 |
0.037 |
| 107 |
253483_at |
AT4G31910
|
BAT1, BR-related AcylTransferase1, PIZ, PIZZA |
-0.245 |
0.024 |
| 108 |
255632_at |
AT4G00680
|
ADF8, actin depolymerizing factor 8 |
-0.237 |
-0.133 |
| 109 |
262756_at |
AT1G16370
|
ATOCT6, ARABIDOPSIS THALIANA ORGANIC CATION/CARNITINE TRANSPORTER 6, OCT6, organic cation/carnitine transporter 6 |
-0.233 |
-0.115 |
| 110 |
258646_at |
AT3G08040
|
ATFRD3, FRD3, FERRIC REDUCTASE DEFECTIVE 3, MAN1, MANGANESE ACCUMULATOR 1 |
-0.232 |
-0.080 |
| 111 |
266941_at |
AT2G18980
|
[Peroxidase superfamily protein] |
-0.225 |
-0.199 |
| 112 |
265897_at |
AT2G25680
|
MOT1, molybdate transporter 1 |
-0.224 |
-0.018 |
| 113 |
251513_at |
AT3G59220
|
ATPIRIN1, PRN, pirin, PRN1 |
-0.221 |
-0.074 |
| 114 |
257203_at |
AT3G23730
|
XTH16, xyloglucan endotransglucosylase/hydrolase 16 |
-0.215 |
-0.049 |
| 115 |
245737_at |
AT1G44160
|
[HSP40/DnaJ peptide-binding protein] |
-0.212 |
0.007 |