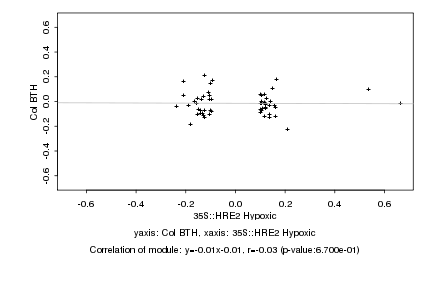
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
35S::HRE2 Hypoxic |
Log2 signal ratio
Col BTH |
| 1 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
0.663 |
-0.015 |
| 2 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.534 |
0.104 |
| 3 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
0.208 |
-0.219 |
| 4 |
249726_at |
AT5G35480
|
unknown |
0.163 |
0.184 |
| 5 |
247252_at |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
0.160 |
-0.114 |
| 6 |
251001_at |
AT5G02670
|
unknown |
0.158 |
-0.044 |
| 7 |
264271_at |
AT1G60270
|
BGLU6, beta glucosidase 6 |
0.155 |
-0.032 |
| 8 |
249542_at |
AT5G38140
|
NF-YC12, nuclear factor Y, subunit C12 |
0.146 |
0.111 |
| 9 |
245440_at |
AT4G16680
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.138 |
0.003 |
| 10 |
263086_at |
AT2G16110
|
[pseudogene] |
0.137 |
-0.099 |
| 11 |
266616_at |
AT2G29680
|
ATCDC6, CDC6, cell division control 6 |
0.137 |
-0.125 |
| 12 |
255219_at |
AT4G07720
|
[pseudogene] |
0.136 |
-0.029 |
| 13 |
255419_at |
AT4G03230
|
[S-locus lectin protein kinase family protein] |
0.123 |
0.026 |
| 14 |
264049_at |
AT2G22390
|
ATRABA4E, RABA4E, RAB GTPase homolog A4E |
0.121 |
-0.050 |
| 15 |
254377_at |
AT4G21650
|
[Subtilase family protein] |
0.121 |
-0.048 |
| 16 |
251228_at |
AT3G62710
|
[Glycosyl hydrolase family protein] |
0.119 |
-0.009 |
| 17 |
254427_at |
AT4G21190
|
emb1417, embryo defective 1417 |
0.119 |
-0.023 |
| 18 |
248364_at |
AT5G52490
|
[Fibrillarin family protein] |
0.116 |
-0.002 |
| 19 |
254304_at |
AT4G22270
|
ATMRB1, ARABIDOPSIS MEMBRANE RELATED BIGGER1, MRB1, MEMBRANE RELATED BIGGER1 |
0.116 |
-0.117 |
| 20 |
264940_at |
AT1G60470
|
AtGolS4, galactinol synthase 4, GolS4, galactinol synthase 4 |
0.114 |
0.064 |
| 21 |
256676_at |
AT3G52180
|
ATPTPKIS1, ATSEX4, DSP4, DUAL-SPECIFICITY PROTEIN PHOSPHATASE 4, SEX4, STARCH-EXCESS 4 |
0.107 |
-0.050 |
| 22 |
260600_at |
AT1G55950
|
[DNA-binding storekeeper protein-related transcriptional regulator] |
0.103 |
0.004 |
| 23 |
248029_at |
AT5G55700
|
BAM4, beta-amylase 4, BMY6, BETA-AMYLASE 6 |
0.102 |
0.054 |
| 24 |
256984_at |
AT3G13480
|
unknown |
0.101 |
-0.072 |
| 25 |
245912_at |
AT5G19600
|
SULTR3;5, sulfate transporter 3;5 |
0.101 |
-0.013 |
| 26 |
251221_at |
AT3G62550
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.100 |
-0.009 |
| 27 |
258874_at |
AT3G03230
|
[alpha/beta-Hydrolases superfamily protein] |
0.099 |
-0.063 |
| 28 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
0.098 |
0.058 |
| 29 |
256418_at |
AT3G06160
|
[AP2/B3-like transcriptional factor family protein] |
0.097 |
-0.088 |
| 30 |
266209_at |
AT2G27550
|
ATC, centroradialis |
-0.240 |
-0.036 |
| 31 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
-0.213 |
0.053 |
| 32 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
-0.211 |
0.164 |
| 33 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
-0.193 |
-0.028 |
| 34 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-0.181 |
-0.180 |
| 35 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
-0.167 |
0.001 |
| 36 |
247657_at |
AT5G59845
|
[Gibberellin-regulated family protein] |
-0.159 |
-0.011 |
| 37 |
246850_at |
AT5G26860
|
LON1, lon protease 1, LON_ARA_ARA |
-0.156 |
0.032 |
| 38 |
247026_at |
AT5G67080
|
MAPKKK19, mitogen-activated protein kinase kinase kinase 19 |
-0.155 |
-0.103 |
| 39 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.151 |
-0.060 |
| 40 |
248048_at |
AT5G56080
|
ATNAS2, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 2, NAS2, nicotianamine synthase 2 |
-0.144 |
-0.065 |
| 41 |
265422_at |
AT2G20800
|
NDB4, NAD(P)H dehydrogenase B4 |
-0.141 |
-0.094 |
| 42 |
255648_at |
AT4G00910
|
[Aluminium activated malate transporter family protein] |
-0.139 |
0.017 |
| 43 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
-0.135 |
-0.089 |
| 44 |
260955_at |
AT1G06000
|
[UDP-Glycosyltransferase superfamily protein] |
-0.130 |
0.042 |
| 45 |
244977_at |
ATCG00730
|
PETD, photosynthetic electron transfer D |
-0.129 |
-0.109 |
| 46 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
-0.128 |
0.214 |
| 47 |
266668_at |
AT2G29760
|
OTP81, ORGANELLE TRANSCRIPT PROCESSING 81 |
-0.127 |
-0.066 |
| 48 |
253191_at |
AT4G35350
|
XCP1, xylem cysteine peptidase 1 |
-0.126 |
-0.127 |
| 49 |
259562_at |
AT1G21200
|
[sequence-specific DNA binding transcription factors] |
-0.110 |
0.078 |
| 50 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-0.107 |
-0.101 |
| 51 |
258996_at |
AT3G01800
|
[Ribosome recycling factor] |
-0.107 |
0.050 |
| 52 |
262081_at |
AT1G59540
|
ZCF125 |
-0.106 |
0.022 |
| 53 |
264809_at |
AT1G08830
|
CSD1, copper/zinc superoxide dismutase 1 |
-0.104 |
0.148 |
| 54 |
258965_at |
AT3G10530
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.103 |
-0.065 |
| 55 |
267463_at |
AT2G33845
|
[Nucleic acid-binding, OB-fold-like protein] |
-0.099 |
0.017 |
| 56 |
264184_at |
AT1G54790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.098 |
-0.078 |
| 57 |
248846_at |
AT5G46500
|
unknown |
-0.095 |
0.170 |