|
probeID |
AGICode |
Annotation |
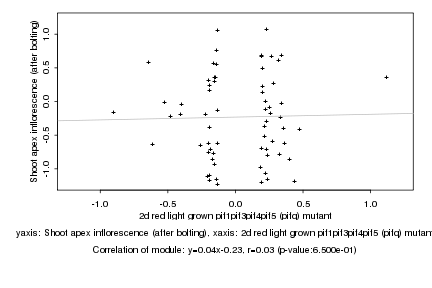
Log2 signal ratio
2d red light grown pif1pif3pif4pif5 (pifq) mutant |
Log2 signal ratio
Shoot apex inflorescence (after bolting) |
| 1 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
1.114 |
0.357 |
| 2 |
245346_at |
AT4G17090
|
AtBAM3, BAM3, BETA-AMYLASE 3, BMY8, BETA-AMYLASE 8, CT-BMY, chloroplast beta-amylase |
0.471 |
-0.409 |
| 3 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
0.431 |
-1.181 |
| 4 |
263098_at |
AT2G16005
|
[MD-2-related lipid recognition domain-containing protein] |
0.396 |
-0.859 |
| 5 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
0.360 |
-0.613 |
| 6 |
253971_at |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
0.350 |
-0.399 |
| 7 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
0.340 |
-0.028 |
| 8 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
0.340 |
0.686 |
| 9 |
245689_at |
AT5G04120
|
[Phosphoglycerate mutase family protein] |
0.332 |
-0.223 |
| 10 |
248728_at |
AT5G48000
|
CYP708 A2, CYTOCHROME P450, FAMILY 708, SUBFAMILY A, POLYPEPTIDE 2, CYP708A2, cytochrome P450, family 708, subfamily A, polypeptide 2, THAH, THALIANOL HYDROXYLASE, THAH1, THALIANOL HYDROXYLASE 1 |
0.320 |
-0.782 |
| 11 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.317 |
0.622 |
| 12 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
0.281 |
0.278 |
| 13 |
246203_at |
AT4G36610
|
[alpha/beta-Hydrolases superfamily protein] |
0.269 |
-0.590 |
| 14 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.264 |
0.676 |
| 15 |
248727_at |
AT5G47990
|
CYP705A5, cytochrome P450, family 705, subfamily A, polypeptide 5, THAD, THALIAN-DIOL DESATURASE, THAD1, THALIAN-DIOL DESATURASE |
0.255 |
-0.167 |
| 16 |
256459_at |
AT1G36180
|
ACC2, acetyl-CoA carboxylase 2 |
0.250 |
-0.088 |
| 17 |
263890_at |
AT2G37030
|
SAUR46, SMALL AUXIN UPREGULATED RNA 46 |
0.230 |
-0.798 |
| 18 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
0.230 |
-1.148 |
| 19 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.229 |
-0.293 |
| 20 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
0.227 |
1.082 |
| 21 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
0.223 |
-0.701 |
| 22 |
255913_at |
AT1G66980
|
GDPDL2, Glycerophosphodiester phosphodiesterase (GDPD) like 2, SNC4, suppressor of npr1-1 constitutive 4 |
0.221 |
-1.061 |
| 23 |
263558_at |
AT2G16380
|
[Sec14p-like phosphatidylinositol transfer family protein] |
0.221 |
0.008 |
| 24 |
254553_at |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
0.220 |
-0.104 |
| 25 |
265511_at |
AT2G05540
|
[Glycine-rich protein family] |
0.210 |
-0.359 |
| 26 |
256603_at |
AT3G28270
|
unknown |
0.208 |
-0.506 |
| 27 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
0.200 |
0.136 |
| 28 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
0.199 |
0.491 |
| 29 |
250239_at |
AT5G13580
|
ABCG6, ATP-binding cassette G6 |
0.196 |
0.235 |
| 30 |
252983_at |
AT4G37980
|
ATCAD7, CAD7, CINNAMYL-ALCOHOL DEHYDROGENASE 7, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-1, elicitor-activated gene 3-1 |
0.192 |
0.697 |
| 31 |
252618_at |
AT3G45140
|
ATLOX2, ARABIODOPSIS THALIANA LIPOXYGENASE 2, LOX2, lipoxygenase 2 |
0.192 |
0.680 |
| 32 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
0.189 |
-1.201 |
| 33 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
0.186 |
-0.684 |
| 34 |
248911_at |
AT5G45830
|
ATDOG1, DOG1, DELAY OF GERMINATION 1, GAAS5, germination ability after storage 5, GSQ5, GLUCOSE SENSING QTL 5 |
0.184 |
-0.966 |
| 35 |
251497_at |
AT3G59060
|
PIF5, PHYTOCHROME-INTERACTING FACTOR 5, PIL6, phytochrome interacting factor 3-like 6 |
-0.907 |
-0.155 |
| 36 |
264510_at |
AT1G09530
|
PAP3, PHYTOCHROME-ASSOCIATED PROTEIN 3, PIF3, phytochrome interacting factor 3, POC1, PHOTOCURRENT 1 |
-0.644 |
0.582 |
| 37 |
265584_at |
AT2G20180
|
PIF1, PHY-INTERACTING FACTOR 1, PIL5, phytochrome interacting factor 3-like 5 |
-0.619 |
-0.624 |
| 38 |
252282_at |
AT3G49360
|
PGL2, 6-phosphogluconolactonase 2 |
-0.525 |
-0.011 |
| 39 |
265248_at |
AT2G43010
|
AtPIF4, PIF4, phytochrome interacting factor 4, SRL2 |
-0.484 |
-0.221 |
| 40 |
254685_at |
AT4G13790
|
SAUR25, SMALL AUXIN UPREGULATED RNA 25 |
-0.409 |
-0.186 |
| 41 |
252763_at |
AT3G42725
|
[Putative membrane lipoprotein] |
-0.405 |
-0.031 |
| 42 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
-0.259 |
-0.647 |
| 43 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.223 |
-0.189 |
| 44 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
-0.213 |
-1.109 |
| 45 |
263690_at |
AT1G26960
|
AtHB23, homeobox protein 23, HB23, homeobox protein 23 |
-0.206 |
0.324 |
| 46 |
259947_at |
AT1G71530
|
[Protein kinase superfamily protein] |
-0.201 |
-0.754 |
| 47 |
257951_at |
AT3G21700
|
ATSGP2, SGP2 |
-0.200 |
-0.616 |
| 48 |
245735_at |
AT1G73410
|
ATMYB54, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 54, MYB54, myb domain protein 54 |
-0.195 |
-1.092 |
| 49 |
249167_at |
AT5G42860
|
unknown |
-0.195 |
0.250 |
| 50 |
253155_at |
AT4G35720
|
unknown |
-0.194 |
-1.173 |
| 51 |
260973_at |
AT1G53490
|
HEI10, homolog of human HEI10 ( Enhancer of cell Invasion No.10) |
-0.194 |
0.168 |
| 52 |
249777_at |
AT5G24210
|
[alpha/beta-Hydrolases superfamily protein] |
-0.192 |
-0.373 |
| 53 |
248197_at |
AT5G54190
|
PORA, protochlorophyllide oxidoreductase A |
-0.185 |
-0.702 |
| 54 |
261772_at |
AT1G76240
|
unknown |
-0.171 |
-0.849 |
| 55 |
245076_at |
AT2G23170
|
GH3.3 |
-0.168 |
0.572 |
| 56 |
254983_at |
AT4G10560
|
MEE53, maternal effect embryo arrest 53 |
-0.166 |
-0.769 |
| 57 |
267409_at |
AT2G34910
|
unknown |
-0.157 |
0.365 |
| 58 |
257418_at |
AT1G30850
|
RSH4, root hair specific 4 |
-0.157 |
-0.935 |
| 59 |
254490_at |
AT4G20320
|
[CTP synthase family protein] |
-0.155 |
0.308 |
| 60 |
245944_at |
AT5G19520
|
ATMSL9, MSL9, mechanosensitive channel of small conductance-like 9 |
-0.151 |
0.363 |
| 61 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.147 |
0.759 |
| 62 |
251046_at |
AT5G02370
|
[ATP binding microtubule motor family protein] |
-0.142 |
0.561 |
| 63 |
248569_at |
AT5G49770
|
[Leucine-rich repeat protein kinase family protein] |
-0.141 |
-1.144 |
| 64 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
-0.137 |
-1.218 |
| 65 |
267006_at |
AT2G34190
|
[Xanthine/uracil permease family protein] |
-0.137 |
1.055 |
| 66 |
251523_at |
AT3G58975
|
[expressed protein] |
-0.136 |
-0.133 |
| 67 |
256518_at |
AT1G66080
|
unknown |
-0.136 |
-0.613 |