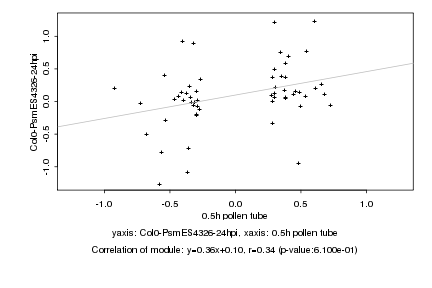
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
0.5h pollen tube |
Log2 signal ratio
Col0-PsmES4326-24hpi |
| 1 |
249794_at |
AT5G23530
|
AtCXE18, carboxyesterase 18, CXE18, carboxyesterase 18 |
0.720 |
-0.053 |
| 2 |
265904_at |
AT2G25630
|
BGLU14, beta glucosidase 14 |
0.678 |
0.120 |
| 3 |
250858_at |
AT5G04760
|
[Duplicated homeodomain-like superfamily protein] |
0.652 |
0.268 |
| 4 |
252045_at |
AT3G52450
|
AtPUB22, PUB22, plant U-box 22 |
0.610 |
0.205 |
| 5 |
259866_at |
AT1G76640
|
CML39, CALMODULIN LIKE 39 |
0.598 |
1.227 |
| 6 |
263935_at |
AT2G35930
|
AtPUB23, PUB23, plant U-box 23 |
0.537 |
0.767 |
| 7 |
259064_at |
AT3G07490
|
AGD11, ARF-GAP domain 11, AtCML3, CML3, calmodulin-like 3 |
0.534 |
0.082 |
| 8 |
258330_at |
AT3G16130
|
ATROPGEF13, PIRF2, phytochrome interacting RopGEF 2, ROPGEF13, ROP (rho of plants) guanine nucleotide exchange factor 13 |
0.491 |
-0.073 |
| 9 |
263469_at |
AT2G31910
|
ATCHX21, cation/H+ exchanger 21, CHX21, cation/H+ exchanger 21 |
0.484 |
0.148 |
| 10 |
254746_at |
AT4G12980
|
[Auxin-responsive family protein] |
0.474 |
-0.940 |
| 11 |
260227_at |
AT1G74450
|
unknown |
0.458 |
0.160 |
| 12 |
267169_at |
AT2G37540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.440 |
0.116 |
| 13 |
256633_at |
AT3G28340
|
GATL10, galacturonosyltransferase-like 10, GolS8, galactinol synthase 8 |
0.398 |
0.701 |
| 14 |
253630_at |
AT4G30490
|
[AFG1-like ATPase family protein] |
0.379 |
0.375 |
| 15 |
260978_at |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
0.378 |
0.586 |
| 16 |
256666_at |
AT3G20670
|
HTA13, histone H2A 13 |
0.376 |
0.058 |
| 17 |
264039_at |
AT2G03740
|
[late embryogenesis abundant domain-containing protein / LEA domain-containing protein] |
0.375 |
0.069 |
| 18 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
0.368 |
0.175 |
| 19 |
247208_at |
AT5G64870
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
0.349 |
0.397 |
| 20 |
254710_at |
AT4G18050
|
ABCB9, ATP-binding cassette B9, PGP9, P-glycoprotein 9 |
0.340 |
0.761 |
| 21 |
247792_at |
AT5G58787
|
[RING/U-box superfamily protein] |
0.302 |
0.219 |
| 22 |
253323_at |
AT4G33920
|
APD5, Arabidopsis Pp2c clade D 5 |
0.297 |
0.492 |
| 23 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.296 |
1.223 |
| 24 |
247857_at |
AT5G58400
|
[Peroxidase superfamily protein] |
0.292 |
0.124 |
| 25 |
265686_at |
AT2G24460
|
unknown |
0.291 |
0.071 |
| 26 |
252278_at |
AT3G49530
|
ANAC062, NAC domain containing protein 62, NAC062, NAC domain containing protein 62, NTL6, NTM1 (NAC WITH TRANSMEMBRANE MOTIF 1)-LIKE 6 |
0.282 |
0.377 |
| 27 |
245026_at |
ATCG00140
|
ATPH |
0.281 |
-0.336 |
| 28 |
246118_at |
AT5G20340
|
BG5, beta-1,3-glucanase 5 |
0.279 |
0.006 |
| 29 |
263175_at |
AT1G05510
|
OBAP1a, Oil Body-Associated Protein 1a |
0.272 |
0.105 |
| 30 |
245803_at |
AT1G47128
|
RD21, responsive to dehydration 21, RD21A, responsive to dehydration 21A |
-0.927 |
0.209 |
| 31 |
259574_at |
AT1G35310
|
MLP168, MLP-like protein 168 |
-0.728 |
-0.027 |
| 32 |
266421_at |
AT2G38540
|
ATLTP1, ARABIDOPSIS THALIANA LIPID TRANSFER PROTEIN 1, LP1, lipid transfer protein 1, LTP1, LIPID TRANSFER PROTEIN 1 |
-0.686 |
-0.493 |
| 33 |
259660_at |
AT1G55260
|
LTPG6, glycosylphosphatidylinositol-anchored lipid protein transfer 6 |
-0.580 |
-1.257 |
| 34 |
248921_at |
AT5G45950
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.571 |
-0.780 |
| 35 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
-0.546 |
0.403 |
| 36 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
-0.537 |
-0.284 |
| 37 |
266927_at |
AT2G45960
|
ATHH2, PIP1;2, NAMED PLASMA MEMBRANE INTRINSIC PROTEIN 1;2, PIP1B, plasma membrane intrinsic protein 1B, TMP-A, TRANSMEMBRANE PROTEIN A |
-0.467 |
0.037 |
| 38 |
267104_at |
AT2G41430
|
CID1, CTC-Interacting Domain 1, ERD15, EARLY RESPONSIVE TO DEHYDRATION 15, LSR1, LIGHT STRESS-REGULATED 1 |
-0.435 |
0.083 |
| 39 |
256521_at |
AT1G66120
|
AAE11, acyl-activating enzyme 11, AtAAE11 |
-0.416 |
0.139 |
| 40 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
-0.408 |
0.925 |
| 41 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.398 |
0.018 |
| 42 |
254555_at |
AT4G19570
|
[Chaperone DnaJ-domain superfamily protein] |
-0.374 |
0.132 |
| 43 |
264371_at |
AT1G12090
|
ELP, extensin-like protein |
-0.366 |
-1.087 |
| 44 |
263709_at |
AT1G09310
|
unknown |
-0.363 |
-0.710 |
| 45 |
255345_at |
AT4G04460
|
[Saposin-like aspartyl protease family protein] |
-0.353 |
0.233 |
| 46 |
246072_at |
AT5G20240
|
PI, PISTILLATA |
-0.346 |
0.064 |
| 47 |
253550_at |
AT4G30960
|
ATCIPK6, CBL-INTERACTING PROTEIN KINASE 6, CIPK6, CBL-INTERACTING PROTEIN KINASE 6, SIP3, SOS3-interacting protein 3, SNRK3.14, SNF1-RELATED PROTEIN KINASE 3.14 |
-0.343 |
-0.004 |
| 48 |
247993_at |
AT5G56130
|
AtTEX1, TEX1, THO3 |
-0.325 |
-0.054 |
| 49 |
248918_at |
AT5G45890
|
AtSAG12, SAG12, senescence-associated gene 12 |
-0.322 |
0.896 |
| 50 |
245006_at |
ATCG00340
|
PSAB |
-0.317 |
-0.007 |
| 51 |
247627_at |
AT5G60360
|
AALP, aleurain-like protease, ALP, aleurain-like protease, SAG2, SENESCENCE ASSOCIATED GENE2 |
-0.301 |
0.167 |
| 52 |
262609_at |
AT1G13930
|
unknown |
-0.300 |
-0.213 |
| 53 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
-0.299 |
-0.193 |
| 54 |
252430_at |
AT3G47470
|
CAB4, LHCA4, light-harvesting chlorophyll-protein complex I subunit A4 |
-0.295 |
-0.073 |
| 55 |
255803_at |
AT4G10170
|
[SNARE-like superfamily protein] |
-0.295 |
0.017 |
| 56 |
260128_at |
AT1G36310
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.280 |
-0.121 |
| 57 |
248193_at |
AT5G54080
|
AtHGO, HGO, homogentisate 1,2-dioxygenase |
-0.267 |
0.345 |