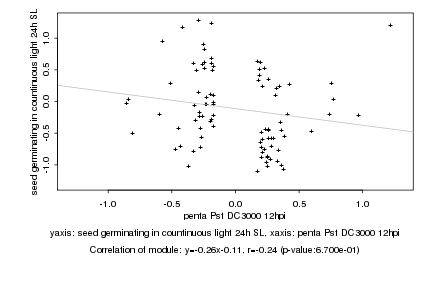
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
penta Pst DC3000 12hpi |
Log2 signal ratio
seed germinating in countinuous light 24h SL |
| 1 |
246432_at |
AT5G17490
|
AtRGL3, RGL3, RGA-like protein 3 |
1.211 |
1.214 |
| 2 |
250063_at |
AT5G17880
|
CSA1, constitutive shade-avoidance1 |
0.962 |
-0.220 |
| 3 |
262010_at |
AT1G35612
|
[expressed protein] |
0.764 |
0.040 |
| 4 |
250064_at |
AT5G17890
|
CHS3, CHILLING SENSITIVE 3, DAR4, DA1-related protein 4 |
0.752 |
0.298 |
| 5 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
0.739 |
-0.196 |
| 6 |
262018_at |
AT1G35617
|
unknown |
0.594 |
-0.466 |
| 7 |
246536_at |
AT5G15920
|
EMB2782, EMBRYO DEFECTIVE 2782, SMC5, structural maintenance of chromosomes 5 |
0.418 |
0.278 |
| 8 |
246498_at |
AT5G16230
|
[Plant stearoyl-acyl-carrier-protein desaturase family protein] |
0.407 |
-0.196 |
| 9 |
256989_at |
AT3G28580
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.382 |
-0.543 |
| 10 |
265705_at |
AT2G03410
|
[Mo25 family protein] |
0.376 |
-1.064 |
| 11 |
248236_at |
AT5G53870
|
AtENODL1, ENODL1, early nodulin-like protein 1 |
0.358 |
-1.008 |
| 12 |
260335_at |
AT1G74000
|
SS3, strictosidine synthase 3 |
0.355 |
-0.449 |
| 13 |
255340_at |
AT4G04490
|
CRK36, cysteine-rich RLK (RECEPTOR-like protein kinase) 36 |
0.353 |
-0.321 |
| 14 |
256833_at |
AT3G22910
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.346 |
0.249 |
| 15 |
251419_at |
AT3G60470
|
unknown |
0.336 |
-0.768 |
| 16 |
250038_at |
AT5G18360
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.324 |
-0.939 |
| 17 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
0.321 |
0.215 |
| 18 |
262666_at |
AT1G14080
|
ATFUT6, FUT6, fucosyltransferase 6 |
0.307 |
0.102 |
| 19 |
259985_at |
AT1G76620
|
unknown |
0.295 |
-0.576 |
| 20 |
260734_at |
AT1G17600
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.281 |
-0.707 |
| 21 |
265030_at |
AT1G61610
|
[S-locus lectin protein kinase family protein] |
0.276 |
-0.570 |
| 22 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
0.269 |
-0.912 |
| 23 |
262813_at |
AT1G11670
|
[MATE efflux family protein] |
0.257 |
0.362 |
| 24 |
265047_at |
AT1G51960
|
IQD27, IQ-domain 27 |
0.256 |
-0.580 |
| 25 |
253696_at |
AT4G29740
|
ATCKX4, CKX4, cytokinin oxidase 4 |
0.256 |
-0.427 |
| 26 |
252014_at |
AT3G52870
|
[IQ calmodulin-binding motif family protein] |
0.253 |
-0.445 |
| 27 |
253259_at |
AT4G34410
|
RRTF1, redox responsive transcription factor 1 |
0.250 |
-0.869 |
| 28 |
262912_at |
AT1G59740
|
AtNPF4.3, NPF4.3, NRT1/ PTR family 4.3 |
0.247 |
-1.019 |
| 29 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
0.247 |
-0.853 |
| 30 |
247913_at |
AT5G57510
|
unknown |
0.244 |
-0.953 |
| 31 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
0.229 |
-0.441 |
| 32 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
0.228 |
-0.746 |
| 33 |
254726_at |
AT4G13660
|
ATPRR2, PRR2, pinoresinol reductase 2 |
0.225 |
0.530 |
| 34 |
245932_at |
AT5G09290
|
[Inositol monophosphatase family protein] |
0.209 |
-0.586 |
| 35 |
258894_at |
AT3G05650
|
AtRLP32, receptor like protein 32, RLP32, receptor like protein 32 |
0.208 |
-0.804 |
| 36 |
260023_at |
AT1G30040
|
ATGA2OX2, gibberellin 2-oxidase, GA2OX2, gibberellin 2-oxidase, GA2OX2, GIBBERELLIN 2-OXIDASE 2 |
0.207 |
0.251 |
| 37 |
252537_at |
AT3G45710
|
[Major facilitator superfamily protein] |
0.202 |
-0.486 |
| 38 |
251970_at |
AT3G53150
|
UGT73D1, UDP-glucosyl transferase 73D1 |
0.202 |
-0.711 |
| 39 |
250024_at |
AT5G18270
|
ANAC087, Arabidopsis NAC domain containing protein 87 |
0.197 |
-0.876 |
| 40 |
255177_at |
AT4G08040
|
ACS11, 1-aminocyclopropane-1-carboxylate synthase 11 |
0.195 |
-0.631 |
| 41 |
260243_at |
AT1G63720
|
unknown |
0.191 |
0.629 |
| 42 |
255485_at |
AT4G02550
|
unknown |
0.189 |
0.513 |
| 43 |
245401_at |
AT4G17670
|
unknown |
0.182 |
0.414 |
| 44 |
245849_at |
AT5G13520
|
[peptidase M1 family protein] |
0.176 |
0.341 |
| 45 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
0.171 |
0.637 |
| 46 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
0.170 |
-1.092 |
| 47 |
255852_at |
AT1G66970
|
GDPDL1, Glycerophosphodiester phosphodiesterase (GDPD) like 1, SVL2, SHV3-like 2 |
-0.861 |
-0.029 |
| 48 |
260141_at |
AT1G66350
|
RGL, RGL1, RGA-like 1 |
-0.841 |
0.034 |
| 49 |
250135_at |
AT5G15360
|
unknown |
-0.812 |
-0.502 |
| 50 |
255912_at |
AT1G66960
|
LUP5 |
-0.597 |
-0.199 |
| 51 |
266331_at |
AT2G01570
|
RGA, REPRESSOR OF GA, RGA1, REPRESSOR OF GA1-3 1 |
-0.581 |
0.948 |
| 52 |
262850_at |
AT1G14920
|
GAI, GIBBERELLIC ACID INSENSITIVE, RGA2, RESTORATION ON GROWTH ON AMMONIA 2 |
-0.512 |
0.286 |
| 53 |
250036_at |
AT5G18340
|
[ARM repeat superfamily protein] |
-0.476 |
-0.756 |
| 54 |
261684_at |
AT1G47400
|
unknown |
-0.449 |
-0.422 |
| 55 |
261375_at |
AT1G53160
|
FTM6, FLORAL TRANSITION AT THE MERISTEM6, SPL4, squamosa promoter binding protein-like 4 |
-0.432 |
-0.697 |
| 56 |
257798_at |
AT3G15950
|
NAI2 |
-0.418 |
1.174 |
| 57 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
-0.370 |
-1.022 |
| 58 |
258792_at |
AT3G04640
|
[glycine-rich protein] |
-0.337 |
0.601 |
| 59 |
247212_at |
AT5G65040
|
unknown |
-0.335 |
-0.782 |
| 60 |
251856_at |
AT3G54720
|
AMP1, ALTERED MERISTEM PROGRAM 1, AtAMP1, COP2, CONSTITUTIVE MORPHOGENESIS 2, HPT, HAUPTLING, MFO1, Multifolia, PT, PRIMORDIA TIMING |
-0.322 |
-0.048 |
| 61 |
250828_at |
AT5G05250
|
unknown |
-0.314 |
-0.297 |
| 62 |
263829_at |
AT2G40435
|
unknown |
-0.311 |
0.501 |
| 63 |
247151_at |
AT5G65640
|
bHLH093, beta HLH protein 93 |
-0.293 |
0.143 |
| 64 |
267472_at |
AT2G02850
|
ARPN, plantacyanin |
-0.291 |
1.294 |
| 65 |
259384_at |
AT3G16450
|
JAL33, Jacalin-related lectin 33 |
-0.288 |
-0.230 |
| 66 |
261607_at |
AT1G49660
|
AtCXE5, carboxyesterase 5, CXE5, carboxyesterase 5 |
-0.286 |
-0.161 |
| 67 |
257365_x_at |
AT2G26020
|
PDF1.2b, plant defensin 1.2b |
-0.282 |
-0.711 |
| 68 |
250100_at |
AT5G16570
|
GLN1;4, glutamine synthetase 1;4 |
-0.280 |
-0.419 |
| 69 |
262232_at |
AT1G68600
|
[Aluminium activated malate transporter family protein] |
-0.271 |
-0.565 |
| 70 |
254125_at |
AT4G24670
|
TAR2, tryptophan aminotransferase related 2 |
-0.262 |
0.594 |
| 71 |
248271_at |
AT5G53420
|
[CCT motif family protein] |
-0.259 |
-0.235 |
| 72 |
249361_at |
AT5G40540
|
[Protein kinase superfamily protein] |
-0.253 |
0.907 |
| 73 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
-0.251 |
0.627 |
| 74 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
-0.249 |
0.529 |
| 75 |
257673_at |
AT3G20370
|
[TRAF-like family protein] |
-0.247 |
0.827 |
| 76 |
258033_at |
AT3G21250
|
ABCC8, ATP-binding cassette C8, ATMRP6, ARABIDOPSIS THALIANA MULTIDRUG RESISTANCE-ASSOCIATED PROTEIN 6, MRP6, multidrug resistance-associated protein 6 |
-0.238 |
-0.045 |
| 77 |
254132_at |
AT4G24660
|
ATHB22, HOMEOBOX PROTEIN 22, HB22, homeobox protein 22, MEE68, MATERNAL EFFECT EMBRYO ARREST 68, ZHD2, ZINC FINGER HOMEODOMAIN 2 |
-0.233 |
0.069 |
| 78 |
253281_at |
AT4G34138
|
UGT73B1, UDP-glucosyl transferase 73B1 |
-0.230 |
-0.045 |
| 79 |
250025_at |
AT5G18290
|
SIP1;2, SIP1B, SMALL AND BASIC INTRINSIC PROTEIN 1B |
-0.203 |
0.120 |
| 80 |
248460_at |
AT5G50915
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.201 |
-0.315 |
| 81 |
251183_at |
AT3G62630
|
unknown |
-0.195 |
0.605 |
| 82 |
258299_at |
AT3G23410
|
ATFAO3, ARABIDOPSIS FATTY ALCOHOL OXIDASE 3, FAO3, fatty alcohol oxidase 3 |
-0.194 |
0.693 |
| 83 |
260876_at |
AT1G21460
|
AtSWEET1, SWEET1 |
-0.191 |
1.246 |
| 84 |
245501_at |
AT4G15620
|
[Uncharacterised protein family (UPF0497)] |
-0.189 |
-0.283 |
| 85 |
250517_at |
AT5G08260
|
scpl35, serine carboxypeptidase-like 35 |
-0.187 |
0.501 |
| 86 |
262170_at |
AT1G74940
|
unknown |
-0.180 |
-0.013 |
| 87 |
257294_at |
AT3G15570
|
[Phototropic-responsive NPH3 family protein] |
-0.179 |
-0.215 |
| 88 |
248306_at |
AT5G52830
|
ATWRKY27, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 27, WRKY27, WRKY DNA-binding protein 27 |
-0.179 |
0.097 |
| 89 |
261708_at |
AT1G32740
|
[SBP (S-ribonuclease binding protein) family protein] |
-0.179 |
-0.386 |
| 90 |
259528_at |
AT1G12330
|
unknown |
-0.178 |
-0.036 |
| 91 |
267481_at |
AT2G02780
|
Leucine-rich repeat protein kinase family protein |
-0.178 |
0.562 |