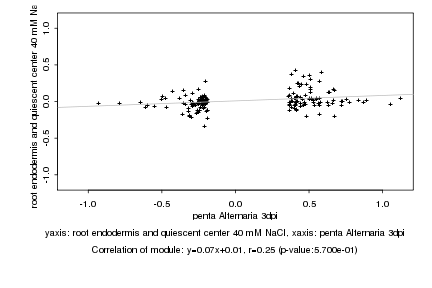
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
penta Alternaria 3dpi |
Log2 signal ratio
root endodermis and quiescent center 40 mM NaCl |
| 1 |
246432_at |
AT5G17490
|
AtRGL3, RGL3, RGA-like protein 3 |
1.119 |
0.043 |
| 2 |
259286_at |
AT3G11480
|
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
1.052 |
-0.029 |
| 3 |
245443_at |
AT4G16730
|
TPS02, terpene synthase 02 |
0.889 |
0.023 |
| 4 |
250063_at |
AT5G17880
|
CSA1, constitutive shade-avoidance1 |
0.870 |
-0.003 |
| 5 |
254574_at |
AT4G19430
|
unknown |
0.834 |
0.026 |
| 6 |
262640_at |
AT1G62760
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.775 |
-0.004 |
| 7 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.748 |
0.030 |
| 8 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
0.721 |
0.003 |
| 9 |
262010_at |
AT1G35612
|
[expressed protein] |
0.717 |
-0.042 |
| 10 |
258182_at |
AT3G21500
|
DXL1, DXS-like 1, DXPS1, 1-deoxy-D-xylulose 5-phosphate synthase 1, DXS2, 1-deoxy-D-xylulose 5-phosphate (DXP) synthase 2 |
0.716 |
0.005 |
| 11 |
251456_at |
AT3G60120
|
BGLU27, beta glucosidase 27 |
0.669 |
0.156 |
| 12 |
245151_at |
AT2G47550
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.667 |
-0.197 |
| 13 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
0.665 |
0.174 |
| 14 |
256349_at |
AT1G54890
|
[Late embryogenesis abundant (LEA) protein-related] |
0.664 |
0.014 |
| 15 |
262018_at |
AT1G35617
|
unknown |
0.653 |
-0.014 |
| 16 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.633 |
0.127 |
| 17 |
267147_at |
AT2G38240
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.632 |
0.135 |
| 18 |
264886_at |
AT1G61120
|
GES, GERANYLLINALOOL SYNTHASE, TPS04, terpene synthase 04, TPS4, TERPENE SYNTHASE 4 |
0.631 |
-0.050 |
| 19 |
250064_at |
AT5G17890
|
CHS3, CHILLING SENSITIVE 3, DAR4, DA1-related protein 4 |
0.624 |
-0.001 |
| 20 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.584 |
0.407 |
| 21 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.573 |
-0.004 |
| 22 |
249454_at |
AT5G39520
|
unknown |
0.571 |
0.053 |
| 23 |
265796_at |
AT2G35730
|
[Heavy metal transport/detoxification superfamily protein ] |
0.570 |
0.283 |
| 24 |
256159_at |
AT1G30135
|
JAZ8, jasmonate-zim-domain protein 8, TIFY5A |
0.569 |
-0.048 |
| 25 |
260608_at |
AT2G43870
|
[Pectin lyase-like superfamily protein] |
0.567 |
-0.173 |
| 26 |
265030_at |
AT1G61610
|
[S-locus lectin protein kinase family protein] |
0.563 |
-0.024 |
| 27 |
267559_at |
AT2G45570
|
CYP76C2, cytochrome P450, family 76, subfamily C, polypeptide 2 |
0.548 |
0.048 |
| 28 |
263363_at |
AT2G03850
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.548 |
0.035 |
| 29 |
265984_at |
AT2G24210
|
TPS10, terpene synthase 10 |
0.534 |
-0.045 |
| 30 |
249860_at |
AT5G22860
|
[Serine carboxypeptidase S28 family protein] |
0.525 |
-0.000 |
| 31 |
265482_at |
AT2G15780
|
[Cupredoxin superfamily protein] |
0.525 |
0.025 |
| 32 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
0.511 |
0.034 |
| 33 |
263073_at |
AT2G17500
|
[Auxin efflux carrier family protein] |
0.511 |
0.047 |
| 34 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.510 |
0.165 |
| 35 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.509 |
0.204 |
| 36 |
259618_at |
AT1G48000
|
AtMYB112, myb domain protein 112, MYB112, myb domain protein 112 |
0.509 |
0.306 |
| 37 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
0.508 |
0.126 |
| 38 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
0.502 |
0.048 |
| 39 |
256024_at |
AT1G58340
|
BCD1, BUSH-AND-CHLOROTIC-DWARF 1, ZF14, ZRZ, ZRIZI |
0.500 |
0.362 |
| 40 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
0.498 |
0.032 |
| 41 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.479 |
0.240 |
| 42 |
259975_at |
AT1G76470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.477 |
-0.203 |
| 43 |
259133_at |
AT3G05400
|
[Major facilitator superfamily protein] |
0.474 |
-0.035 |
| 44 |
260581_at |
AT2G47190
|
ATMYB2, MYB DOMAIN PROTEIN 2, MYB2, myb domain protein 2 |
0.471 |
0.087 |
| 45 |
254101_at |
AT4G25000
|
AMY1, alpha-amylase-like, ATAMY1 |
0.465 |
-0.004 |
| 46 |
249527_at |
AT5G38710
|
[Methylenetetrahydrofolate reductase family protein] |
0.464 |
-0.027 |
| 47 |
259036_at |
AT3G09220
|
LAC7, laccase 7 |
0.462 |
-0.049 |
| 48 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.456 |
0.343 |
| 49 |
254598_at |
AT4G18990
|
XTH29, xyloglucan endotransglucosylase/hydrolase 29 |
0.452 |
0.041 |
| 50 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
0.448 |
0.239 |
| 51 |
249154_at |
AT5G43410
|
[Integrase-type DNA-binding superfamily protein] |
0.442 |
-0.019 |
| 52 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
0.441 |
0.061 |
| 53 |
262666_at |
AT1G14080
|
ATFUT6, FUT6, fucosyltransferase 6 |
0.436 |
-0.059 |
| 54 |
260386_at |
AT1G74010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.431 |
0.214 |
| 55 |
259445_at |
AT1G02400
|
ATGA2OX4, Arabidopsis thaliana gibberellin 2-oxidase 4, ATGA2OX6, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 6, DTA1, DOWNSTREAM TARGET OF AGL15 1, GA2OX6, gibberellin 2-oxidase 6 |
0.424 |
0.257 |
| 56 |
259439_at |
AT1G01480
|
ACS2, 1-amino-cyclopropane-1-carboxylate synthase 2, AT-ACC2 |
0.419 |
0.011 |
| 57 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
0.417 |
0.077 |
| 58 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.415 |
0.246 |
| 59 |
251293_at |
AT3G61930
|
unknown |
0.414 |
-0.111 |
| 60 |
246536_at |
AT5G15920
|
EMB2782, EMBRYO DEFECTIVE 2782, SMC5, structural maintenance of chromosomes 5 |
0.411 |
0.073 |
| 61 |
254597_at |
AT4G18980
|
AtS40-3, AtS40-3 |
0.410 |
0.006 |
| 62 |
251019_at |
AT5G02420
|
unknown |
0.409 |
-0.108 |
| 63 |
261242_at |
AT1G32960
|
ATSBT3.3, SBT3.3 |
0.409 |
-0.004 |
| 64 |
267389_at |
AT2G44460
|
BGLU28, beta glucosidase 28 |
0.408 |
0.429 |
| 65 |
260023_at |
AT1G30040
|
ATGA2OX2, gibberellin 2-oxidase, GA2OX2, gibberellin 2-oxidase, GA2OX2, GIBBERELLIN 2-OXIDASE 2 |
0.407 |
-0.035 |
| 66 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
0.406 |
-0.012 |
| 67 |
248528_at |
AT5G50760
|
SAUR55, SMALL AUXIN UPREGULATED RNA 55 |
0.405 |
0.043 |
| 68 |
261150_at |
AT1G19640
|
JMT, jasmonic acid carboxyl methyltransferase |
0.405 |
-0.042 |
| 69 |
250292_at |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
0.400 |
-0.042 |
| 70 |
251743_at |
AT3G55890
|
[Yippee family putative zinc-binding protein] |
0.398 |
-0.085 |
| 71 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.395 |
0.046 |
| 72 |
265913_at |
AT2G25625
|
unknown |
0.390 |
0.122 |
| 73 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
0.384 |
0.008 |
| 74 |
261394_at |
AT1G79680
|
ATWAKL10, WAKL10, WALL ASSOCIATED KINASE (WAK)-LIKE 10 |
0.380 |
0.006 |
| 75 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.377 |
0.373 |
| 76 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
0.376 |
-0.071 |
| 77 |
252334_at |
AT3G48850
|
MPT2, mitochondrial phosphate transporter 2, PHT3;2, phosphate transporter 3;2 |
0.375 |
0.042 |
| 78 |
260933_at |
AT1G02470
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.372 |
-0.003 |
| 79 |
251766_at |
AT3G55910
|
unknown |
0.367 |
-0.009 |
| 80 |
260335_at |
AT1G74000
|
SS3, strictosidine synthase 3 |
0.364 |
0.189 |
| 81 |
262133_at |
AT1G78000
|
SEL1, SELENATE RESISTANT 1, SULTR1;2, sulfate transporter 1;2 |
0.362 |
-0.031 |
| 82 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
0.362 |
-0.111 |
| 83 |
263680_at |
AT1G26930
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.361 |
-0.047 |
| 84 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
0.361 |
0.089 |
| 85 |
249057_at |
AT5G44480
|
DUR, DEFECTIVE UGE IN ROOT |
0.360 |
0.079 |
| 86 |
255852_at |
AT1G66970
|
GDPDL1, Glycerophosphodiester phosphodiesterase (GDPD) like 1, SVL2, SHV3-like 2 |
-0.934 |
-0.018 |
| 87 |
250135_at |
AT5G15360
|
unknown |
-0.788 |
-0.020 |
| 88 |
260141_at |
AT1G66350
|
RGL, RGL1, RGA-like 1 |
-0.647 |
-0.002 |
| 89 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.614 |
-0.068 |
| 90 |
262850_at |
AT1G14920
|
GAI, GIBBERELLIC ACID INSENSITIVE, RGA2, RESTORATION ON GROWTH ON AMMONIA 2 |
-0.599 |
-0.042 |
| 91 |
266331_at |
AT2G01570
|
RGA, REPRESSOR OF GA, RGA1, REPRESSOR OF GA1-3 1 |
-0.552 |
-0.065 |
| 92 |
261684_at |
AT1G47400
|
unknown |
-0.505 |
0.035 |
| 93 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-0.497 |
0.070 |
| 94 |
261375_at |
AT1G53160
|
FTM6, FLORAL TRANSITION AT THE MERISTEM6, SPL4, squamosa promoter binding protein-like 4 |
-0.479 |
0.046 |
| 95 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
-0.470 |
-0.078 |
| 96 |
245965_at |
AT5G19730
|
[Pectin lyase-like superfamily protein] |
-0.431 |
0.138 |
| 97 |
254794_at |
AT4G12970
|
EPFL9, STOMAGEN, STOMAGEN |
-0.385 |
0.049 |
| 98 |
246550_at |
AT5G14920
|
GASA14, A-stimulated in Arabidopsis 14 |
-0.364 |
-0.174 |
| 99 |
248921_at |
AT5G45950
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.357 |
0.161 |
| 100 |
245980_at |
AT5G13140
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.356 |
-0.015 |
| 101 |
250036_at |
AT5G18340
|
[ARM repeat superfamily protein] |
-0.346 |
0.095 |
| 102 |
260041_at |
AT1G68780
|
[RNI-like superfamily protein] |
-0.343 |
-0.030 |
| 103 |
259009_at |
AT3G09260
|
BGLU23, LEB, LONG ER BODY, PSR3.1, PYK10 |
-0.324 |
-0.092 |
| 104 |
252943_at |
AT4G39330
|
ATCAD9, CAD9, cinnamyl alcohol dehydrogenase 9 |
-0.322 |
-0.126 |
| 105 |
250828_at |
AT5G05250
|
unknown |
-0.317 |
-0.184 |
| 106 |
265405_at |
AT2G16750
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.314 |
-0.193 |
| 107 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
-0.310 |
0.018 |
| 108 |
261949_at |
AT1G64670
|
BDG1, BODYGUARD1, CED1, 9-CIS EPOXYCAROTENOID DIOXYGENASE DEFECTIVE 1 |
-0.305 |
-0.027 |
| 109 |
245612_at |
AT4G14440
|
ATECI3, ARABIDOPSIS THALIANA DELTA(3), DELTA(2)-ENOYL COA ISOMERASE 3, ECI3, DELTA(3), DELTA(2)-ENOYL COA ISOMERASE 3, HCD1, 3-hydroxyacyl-CoA dehydratase 1 |
-0.304 |
-0.206 |
| 110 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
-0.298 |
-0.016 |
| 111 |
252215_at |
AT3G50240
|
KICP-02 |
-0.297 |
-0.000 |
| 112 |
254293_at |
AT4G23060
|
IQD22, IQ-domain 22 |
-0.293 |
0.120 |
| 113 |
259786_at |
AT1G29660
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.292 |
-0.056 |
| 114 |
249278_at |
AT5G41900
|
[alpha/beta-Hydrolases superfamily protein] |
-0.291 |
-0.028 |
| 115 |
250980_at |
AT5G03130
|
unknown |
-0.286 |
-0.021 |
| 116 |
260982_at |
AT1G53520
|
AtFAP3, FAP3, fatty-acid-binding protein 3 |
-0.277 |
-0.049 |
| 117 |
259592_at |
AT1G27950
|
LTPG1, glycosylphosphatidylinositol-anchored lipid protein transfer 1 |
-0.265 |
-0.162 |
| 118 |
255732_at |
AT1G25450
|
CER60, ECERIFERUM 60, KCS5, 3-ketoacyl-CoA synthase 5 |
-0.263 |
-0.145 |
| 119 |
248784_at |
AT5G47380
|
unknown |
-0.263 |
-0.016 |
| 120 |
264611_at |
AT1G04680
|
[Pectin lyase-like superfamily protein] |
-0.263 |
-0.119 |
| 121 |
262414_at |
AT1G49430
|
LACS2, long-chain acyl-CoA synthetase 2, LRD2, LATERAL ROOT DEVELOPMENT 2 |
-0.261 |
-0.003 |
| 122 |
257547_at |
AT3G13000
|
unknown |
-0.257 |
0.177 |
| 123 |
259375_at |
AT3G16370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.253 |
-0.137 |
| 124 |
267078_at |
AT2G40960
|
[Single-stranded nucleic acid binding R3H protein] |
-0.253 |
0.039 |
| 125 |
266988_at |
AT2G39310
|
JAL22, jacalin-related lectin 22 |
-0.252 |
-0.034 |
| 126 |
255294_at |
AT4G04750
|
[Major facilitator superfamily protein] |
-0.252 |
-0.025 |
| 127 |
265722_at |
AT2G40100
|
LHCB4.3, light harvesting complex photosystem II |
-0.251 |
0.004 |
| 128 |
252711_at |
AT3G43720
|
LTPG2, glycosylphosphatidylinositol-anchored lipid protein transfer 2 |
-0.250 |
-0.113 |
| 129 |
251586_at |
AT3G58070
|
GIS, GLABROUS INFLORESCENCE STEMS |
-0.249 |
0.015 |
| 130 |
267481_at |
AT2G02780
|
Leucine-rich repeat protein kinase family protein |
-0.249 |
-0.120 |
| 131 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
-0.241 |
0.024 |
| 132 |
267497_at |
AT2G30540
|
[Thioredoxin superfamily protein] |
-0.240 |
0.055 |
| 133 |
255817_at |
AT2G33330
|
PDLP3, plasmodesmata-located protein 3 |
-0.239 |
0.068 |
| 134 |
250696_at |
AT5G06790
|
unknown |
-0.238 |
-0.069 |
| 135 |
251174_at |
AT3G63200
|
PLA IIIB, PLP9, PATATIN-like protein 9 |
-0.234 |
0.026 |
| 136 |
261981_at |
AT1G33811
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.233 |
0.038 |
| 137 |
262538_at |
AT1G17140
|
ICR1, interactor of constitutive active rops 1, RIP1, ROP INTERACTIVE PARTNER 1 |
-0.231 |
0.061 |
| 138 |
259760_at |
AT1G77580
|
unknown |
-0.231 |
-0.021 |
| 139 |
259308_at |
AT3G05180
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.227 |
-0.034 |
| 140 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
-0.226 |
0.031 |
| 141 |
257137_at |
AT3G28860
|
ABCB19, ATP-binding cassette B19, ATABCB19, Arabidopsis thaliana ATP-binding cassette B19, ATMDR1, ATMDR11, MULTIDRUG RESISTANCE PROTEIN 11, ATPGP19, MDR1, MDR11, PGP19, P-GLYCOPROTEIN 19 |
-0.226 |
-0.001 |
| 142 |
246919_at |
AT5G25460
|
DGR2, DUF642 L-GalL responsive gene 2 |
-0.225 |
-0.045 |
| 143 |
260685_at |
AT1G17650
|
AtGLYR2, GLYR2, glyoxylate reductase 2, GR2, GLYOXYLATE REDUCTASE 2 |
-0.225 |
0.069 |
| 144 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.222 |
0.026 |
| 145 |
264782_at |
AT1G08810
|
AtMYB60, myb domain protein 60, MYB60, myb domain protein 60 |
-0.221 |
0.020 |
| 146 |
252971_at |
AT4G38770
|
ATPRP4, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 4, PRP4, proline-rich protein 4 |
-0.221 |
0.035 |
| 147 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
-0.219 |
0.055 |
| 148 |
261421_at |
AT1G18840
|
IQD30, IQ-domain 30 |
-0.218 |
-0.089 |
| 149 |
250642_at |
AT5G07180
|
ERL2, ERECTA-like 2 |
-0.217 |
-0.009 |
| 150 |
245112_at |
AT2G41540
|
GPDHC1 |
-0.217 |
0.084 |
| 151 |
247288_at |
AT5G64330
|
JK218, NPH3, NON-PHOTOTROPIC HYPOCOTYL 3, RPT3, ROOT PHOTOTROPISM 3 |
-0.217 |
-0.329 |
| 152 |
261593_at |
AT1G33170
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.216 |
-0.004 |
| 153 |
262830_at |
AT1G14700
|
ATPAP3, PAP3, purple acid phosphatase 3 |
-0.216 |
0.070 |
| 154 |
249288_at |
AT5G41050
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.212 |
-0.015 |
| 155 |
253286_at |
AT4G34260
|
AXY8, ALTERED XYLOGLUCAN 8, FUC95A |
-0.211 |
-0.072 |
| 156 |
259173_at |
AT3G03640
|
BGLU25, beta glucosidase 25, GLUC |
-0.209 |
0.278 |
| 157 |
248420_at |
AT5G51560
|
[Leucine-rich repeat protein kinase family protein] |
-0.208 |
-0.039 |
| 158 |
257275_at |
AT3G14450
|
CID9, CTC-interacting domain 9 |
-0.206 |
0.063 |
| 159 |
264697_at |
AT1G70210
|
ATCYCD1;1, CYCD1;1, CYCLIN D1;1 |
-0.205 |
0.018 |
| 160 |
245616_at |
AT4G14480
|
[Protein kinase superfamily protein] |
-0.205 |
0.021 |
| 161 |
266421_at |
AT2G38540
|
ATLTP1, ARABIDOPSIS THALIANA LIPID TRANSFER PROTEIN 1, LP1, lipid transfer protein 1, LTP1, LIPID TRANSFER PROTEIN 1 |
-0.203 |
-0.135 |
| 162 |
257750_at |
AT3G18800
|
unknown |
-0.202 |
0.017 |
| 163 |
262969_at |
AT1G75710
|
[C2H2-like zinc finger protein] |
-0.202 |
0.053 |
| 164 |
249060_at |
AT5G44560
|
VPS2.2 |
-0.202 |
-0.013 |
| 165 |
247100_at |
AT5G66520
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.199 |
0.001 |
| 166 |
262680_at |
AT1G75880
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.195 |
-0.012 |
| 167 |
246557_at |
AT5G15510
|
[TPX2 (targeting protein for Xklp2) protein family] |
-0.195 |
-0.219 |
| 168 |
246952_at |
AT5G04820
|
ATOFP13, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 13, OFP13, ovate family protein 13 |
-0.195 |
-0.116 |
| 169 |
259832_at |
AT1G69580
|
[Homeodomain-like superfamily protein] |
-0.194 |
-0.114 |
| 170 |
260081_at |
AT1G78170
|
unknown |
-0.193 |
0.028 |