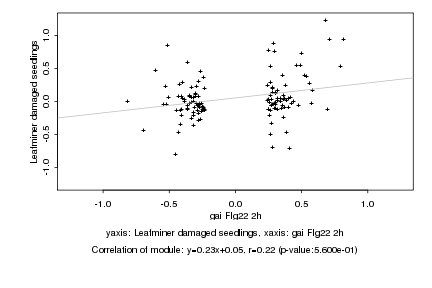
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
gai Flg22 2h |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.812 |
0.951 |
| 2 |
254234_at |
AT4G23680
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.787 |
0.538 |
| 3 |
259866_at |
AT1G76640
|
CML39, CALMODULIN LIKE 39 |
0.707 |
0.956 |
| 4 |
254452_at |
AT4G21100
|
DDB1B, damaged DNA binding protein 1B |
0.691 |
-0.120 |
| 5 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
0.673 |
1.244 |
| 6 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
0.582 |
0.168 |
| 7 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
0.570 |
-0.025 |
| 8 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
0.554 |
0.285 |
| 9 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
0.530 |
0.384 |
| 10 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
0.521 |
0.405 |
| 11 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.496 |
0.732 |
| 12 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
0.490 |
0.551 |
| 13 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
0.474 |
-0.050 |
| 14 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
0.455 |
0.558 |
| 15 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
0.436 |
0.015 |
| 16 |
258923_at |
AT3G10450
|
SCPL7, serine carboxypeptidase-like 7 |
0.417 |
-0.027 |
| 17 |
250464_at |
AT5G10040
|
unknown |
0.413 |
0.076 |
| 18 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
0.404 |
-0.712 |
| 19 |
250550_at |
AT5G07870
|
[HXXXD-type acyl-transferase family protein] |
0.400 |
-0.079 |
| 20 |
252168_at |
AT3G50440
|
ATMES10, ARABIDOPSIS THALIANA METHYL ESTERASE 10, MES10, methyl esterase 10 |
0.399 |
0.048 |
| 21 |
254451_at |
AT4G21090
|
ATMFDX2, ARABIDOPSIS MITOCHONDRIAL FERREDOXIN 2, MFDX2, MITOCHONDRIAL FERREDOXIN 2 |
0.385 |
0.017 |
| 22 |
255774_at |
AT1G18620
|
TRM3, TON1 Recruiting Motif 3 |
0.385 |
-0.463 |
| 23 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.373 |
0.244 |
| 24 |
263151_at |
AT1G54120
|
unknown |
0.370 |
-0.090 |
| 25 |
264507_at |
AT1G09415
|
NIMIN-3, NIM1-interacting 3 |
0.368 |
0.052 |
| 26 |
246493_at |
AT5G16180
|
ATCRS1, ARABIDOPSIS ORTHOLOG OF MAIZE CHLOROPLAST SPLICING FACTOR CRS1, CRS1, ortholog of maize chloroplast splicing factor CRS1 |
0.363 |
-0.237 |
| 27 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.358 |
0.097 |
| 28 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
0.357 |
0.040 |
| 29 |
260662_at |
AT1G19540
|
[NmrA-like negative transcriptional regulator family protein] |
0.355 |
0.406 |
| 30 |
251169_at |
AT3G63210
|
MARD1, MEDIATOR OF ABA-REGULATED DORMANCY 1 |
0.348 |
-0.050 |
| 31 |
256818_at |
AT3G21420
|
LBO1, LATERAL BRANCHING OXIDOREDUCTASE 1 |
0.342 |
-0.099 |
| 32 |
250844_at |
AT5G04470
|
SIM, SIAMESE |
0.339 |
0.049 |
| 33 |
261567_at |
AT1G33055
|
unknown |
0.335 |
0.015 |
| 34 |
254027_at |
AT4G25835
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.314 |
0.060 |
| 35 |
261086_at |
AT1G17460
|
TRFL3, TRF-like 3 |
0.314 |
-0.120 |
| 36 |
266392_at |
AT2G41280
|
ATM10, M10 |
0.312 |
0.024 |
| 37 |
264902_at |
AT1G23060
|
MDP40, MICROTUBULE DESTABILIZING PROTEIN 40 |
0.311 |
0.177 |
| 38 |
250766_at |
AT5G05550
|
[sequence-specific DNA binding transcription factors] |
0.302 |
-0.099 |
| 39 |
247005_at |
AT5G67520
|
adenosine-5'-phosphosulfate (APS) kinase 4 |
0.299 |
0.149 |
| 40 |
251986_at |
AT3G53310
|
[AP2/B3-like transcriptional factor family protein] |
0.299 |
0.122 |
| 41 |
258110_at |
AT3G14610
|
CYP72A7, cytochrome P450, family 72, subfamily A, polypeptide 7 |
0.298 |
-0.006 |
| 42 |
259210_at |
AT3G09150
|
ATHY2, ARABIDOPSIS ELONGATED HYPOCOTYL 2, GUN3, GENOMES UNCOUPLED 3, HY2, ELONGATED HYPOCOTYL 2 |
0.298 |
-0.033 |
| 43 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.295 |
0.773 |
| 44 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
0.294 |
-0.017 |
| 45 |
253285_at |
AT4G34250
|
KCS16, 3-ketoacyl-CoA synthase 16 |
0.289 |
-0.103 |
| 46 |
266271_at |
AT2G29440
|
ATGSTU6, glutathione S-transferase tau 6, GST24, GLUTATHIONE S-TRANSFERASE 24, GSTU6, glutathione S-transferase tau 6 |
0.284 |
0.889 |
| 47 |
247924_at |
AT5G57655
|
[xylose isomerase family protein] |
0.279 |
0.149 |
| 48 |
256061_at |
AT1G07040
|
unknown |
0.277 |
0.220 |
| 49 |
251235_at |
AT3G62860
|
[alpha/beta-Hydrolases superfamily protein] |
0.277 |
0.205 |
| 50 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
0.276 |
-0.689 |
| 51 |
264508_at |
AT1G09570
|
FHY2, FAR RED ELONGATED HYPOCOTYL 2, FRE1, FAR RED ELONGATED 1, HY8, ELONGATED HYPOCOTYL 8, PHYA, phytochrome A |
0.275 |
-0.034 |
| 52 |
249999_at |
AT5G18640
|
[alpha/beta-Hydrolases superfamily protein] |
0.270 |
-0.050 |
| 53 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
0.269 |
0.039 |
| 54 |
258227_at |
AT3G15620
|
UVR3, UV REPAIR DEFECTIVE 3 |
0.266 |
-0.052 |
| 55 |
266324_at |
AT2G46710
|
ROPGAP3, ROP guanosine triphosphatase (GTPase)-activating protein 3 |
0.265 |
-0.327 |
| 56 |
250032_at |
AT5G18170
|
GDH1, glutamate dehydrogenase 1 |
0.264 |
0.297 |
| 57 |
256965_at |
AT3G13450
|
DIN4, DARK INDUCIBLE 4 |
0.262 |
0.097 |
| 58 |
267078_at |
AT2G40960
|
[Single-stranded nucleic acid binding R3H protein] |
0.261 |
-0.132 |
| 59 |
265584_at |
AT2G20180
|
PIF1, PHY-INTERACTING FACTOR 1, PIL5, phytochrome interacting factor 3-like 5 |
0.260 |
-0.488 |
| 60 |
245734_at |
AT1G73480
|
[alpha/beta-Hydrolases superfamily protein] |
0.259 |
0.543 |
| 61 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
0.253 |
-0.208 |
| 62 |
258405_at |
AT3G17590
|
BSH, BUSHY GROWTH, CHE1 |
0.250 |
-0.001 |
| 63 |
262322_at |
AT1G27590
|
unknown |
0.247 |
-0.109 |
| 64 |
265216_at |
AT1G05100
|
MAPKKK18, mitogen-activated protein kinase kinase kinase 18 |
0.246 |
0.778 |
| 65 |
245831_at |
AT1G48840
|
unknown |
0.245 |
0.035 |
| 66 |
267532_at |
AT2G42040
|
unknown |
0.245 |
-0.109 |
| 67 |
258434_at |
AT3G16770
|
ATEBP, ethylene-responsive element binding protein, EBP, ethylene-responsive element binding protein, ERF72, ETHYLENE RESPONSE FACTOR 72, RAP2.3, RELATED TO AP2 3 |
0.239 |
0.020 |
| 68 |
259015_at |
AT3G07350
|
unknown |
0.238 |
0.249 |
| 69 |
266784_at |
AT2G28960
|
[Leucine-rich repeat protein kinase family protein] |
-0.820 |
0.013 |
| 70 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
-0.695 |
-0.434 |
| 71 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
-0.605 |
0.472 |
| 72 |
266785_at |
AT2G28970
|
[Leucine-rich repeat protein kinase family protein] |
-0.548 |
-0.042 |
| 73 |
249051_at |
AT5G44390
|
[FAD-binding Berberine family protein] |
-0.534 |
0.243 |
| 74 |
262905_at |
AT1G59730
|
ATH7, thioredoxin H-type 7, TH7, thioredoxin H-type 7 |
-0.523 |
-0.044 |
| 75 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
-0.517 |
0.858 |
| 76 |
258287_at |
AT3G15990
|
SULTR3;4, sulfate transporter 3;4 |
-0.506 |
0.076 |
| 77 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
-0.460 |
-0.796 |
| 78 |
248321_at |
AT5G52740
|
[Copper transport protein family] |
-0.447 |
-0.133 |
| 79 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
-0.436 |
-0.461 |
| 80 |
249167_at |
AT5G42860
|
unknown |
-0.434 |
0.083 |
| 81 |
246373_at |
AT1G51860
|
[Leucine-rich repeat protein kinase family protein] |
-0.423 |
0.266 |
| 82 |
248877_at |
AT5G46140
|
unknown |
-0.421 |
-0.135 |
| 83 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.421 |
-0.348 |
| 84 |
255302_at |
AT4G04830
|
ATMSRB5, methionine sulfoxide reductase B5, MSRB5, methionine sulfoxide reductase B5 |
-0.415 |
0.085 |
| 85 |
250955_at |
AT5G03190
|
CPUORF47, conserved peptide upstream open reading frame 47 |
-0.415 |
-0.120 |
| 86 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
-0.414 |
-0.212 |
| 87 |
266669_at |
AT2G29750
|
UGT71C1, UDP-glucosyl transferase 71C1 |
-0.406 |
0.047 |
| 88 |
262444_at |
AT1G47480
|
[alpha/beta-Hydrolases superfamily protein] |
-0.404 |
0.292 |
| 89 |
245300_at |
AT4G16350
|
CBL6, calcineurin B-like protein 6, SCABP2, SOS3-LIKE CALCIUM BINDING PROTEIN 2 |
-0.391 |
0.008 |
| 90 |
244901_at |
ATMG00640
|
ORF25 |
-0.385 |
0.044 |
| 91 |
260867_at |
AT1G43790
|
TED6, tracheary element differentiation-related 6 |
-0.369 |
-0.105 |
| 92 |
262357_at |
AT1G73040
|
[Mannose-binding lectin superfamily protein] |
-0.368 |
-0.121 |
| 93 |
249097_at |
AT5G43520
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.365 |
-0.049 |
| 94 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
-0.363 |
0.603 |
| 95 |
249435_at |
AT5G39970
|
[catalytics] |
-0.351 |
-0.029 |
| 96 |
254971_at |
AT4G10380
|
AtNIP5;1, NIP5;1, NOD26-like intrinsic protein 5;1, NLM6, NOD26-LIKE MIP 6, NLM8, NOD26-LIKE MIP 8 |
-0.349 |
0.090 |
| 97 |
259149_at |
AT3G10340
|
PAL4, phenylalanine ammonia-lyase 4 |
-0.349 |
0.101 |
| 98 |
264527_at |
AT1G30760
|
[FAD-binding Berberine family protein] |
-0.339 |
0.220 |
| 99 |
248829_at |
AT5G47130
|
[Bax inhibitor-1 family protein] |
-0.338 |
-0.002 |
| 100 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
-0.336 |
0.073 |
| 101 |
262646_at |
AT1G62800
|
ASP4, aspartate aminotransferase 4 |
-0.333 |
-0.246 |
| 102 |
245080_at |
AT2G23300
|
[Leucine-rich repeat protein kinase family protein] |
-0.324 |
0.013 |
| 103 |
266325_at |
AT2G46630
|
unknown |
-0.324 |
-0.361 |
| 104 |
262615_at |
AT1G13950
|
ATELF5A-1, EUKARYOTIC ELONGATION FACTOR 5A-1, EIF-5A, EIF5A, EUKARYOTIC ELONGATION FACTOR 5A, ELF5A-1, eukaryotic elongation factor 5A-1 |
-0.323 |
0.064 |
| 105 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
-0.320 |
-0.157 |
| 106 |
245302_at |
AT4G17695
|
KAN3, KANADI 3 |
-0.320 |
-0.208 |
| 107 |
266309_at |
AT2G27140
|
[HSP20-like chaperones superfamily protein] |
-0.316 |
-0.079 |
| 108 |
246912_at |
AT5G25820
|
[Exostosin family protein] |
-0.308 |
0.082 |
| 109 |
267588_at |
AT2G42060
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.308 |
0.131 |
| 110 |
248909_at |
AT5G45810
|
CIPK19, CBL-interacting protein kinase 19, SnRK3.5, SNF1-RELATED PROTEIN KINASE 3.5 |
-0.302 |
0.110 |
| 111 |
250646_at |
AT5G06720
|
ATPA2, peroxidase 2, AtPRX53, PA2, peroxidase 2, PRX53, peroxidase 53 |
-0.299 |
0.230 |
| 112 |
264906_at |
AT2G17270
|
MPT1, mitochondrial phosphate transporter 1, PHT3;3, phosphate transporter 3;3 |
-0.296 |
-0.020 |
| 113 |
262533_at |
AT1G17090
|
unknown |
-0.292 |
-0.136 |
| 114 |
244994_at |
ATCG01010
|
NDHF |
-0.289 |
-0.052 |
| 115 |
260039_at |
AT1G68795
|
CLE12, CLAVATA3/ESR-RELATED 12 |
-0.284 |
0.313 |
| 116 |
260890_at |
AT1G29090
|
[Cysteine proteinases superfamily protein] |
-0.284 |
-0.046 |
| 117 |
267590_at |
AT2G39700
|
ATEXP4, ATEXPA4, expansin A4, ATHEXP ALPHA 1.6, EXPA4, expansin A4 |
-0.283 |
0.079 |
| 118 |
253191_at |
AT4G35350
|
XCP1, xylem cysteine peptidase 1 |
-0.282 |
-0.175 |
| 119 |
267094_at |
AT2G38080
|
ATLMCO4, ARABIDOPSIS LACCASE-LIKE MULTICOPPER OXIDASE 4, IRX12, IRREGULAR XYLEM 12, LAC4, LACCASE 4, LMCO4, LACCASE-LIKE MULTICOPPER OXIDASE 4 |
-0.280 |
-0.273 |
| 120 |
251918_at |
AT3G54040
|
[PAR1 protein] |
-0.277 |
-0.025 |
| 121 |
252457_at |
AT3G47180
|
[RING/U-box superfamily protein] |
-0.275 |
-0.040 |
| 122 |
245891_at |
AT5G09220
|
AAP2, amino acid permease 2 |
-0.273 |
-0.083 |
| 123 |
265660_at |
AT2G25470
|
AtRLP21, receptor like protein 21, RLP21, receptor like protein 21 |
-0.268 |
0.467 |
| 124 |
257715_at |
AT3G12750
|
AtZIP1, ZIP1, zinc transporter 1 precursor |
-0.265 |
-0.266 |
| 125 |
251297_at |
AT3G62020
|
GLP10, germin-like protein 10 |
-0.261 |
-0.141 |
| 126 |
262713_at |
AT1G16520
|
unknown |
-0.257 |
-0.147 |
| 127 |
262827_at |
AT1G13100
|
CYP71B29, cytochrome P450, family 71, subfamily B, polypeptide 29 |
-0.257 |
-0.023 |
| 128 |
257600_at |
AT3G24770
|
CLE41, CLAVATA3/ESR-RELATED 41 |
-0.256 |
-0.107 |
| 129 |
266166_at |
AT2G28080
|
[UDP-Glycosyltransferase superfamily protein] |
-0.254 |
-0.063 |
| 130 |
257173_at |
AT3G23810
|
ATSAHH2, S-ADENOSYL-L-HOMOCYSTEINE (SAH) HYDROLASE 2, SAHH2, S-adenosyl-l-homocysteine (SAH) hydrolase 2 |
-0.245 |
-0.097 |
| 131 |
257375_at |
AT2G38640
|
unknown |
-0.243 |
0.377 |
| 132 |
245002_at |
ATCG00270
|
PSBD, photosystem II reaction center protein D |
-0.242 |
-0.081 |
| 133 |
254402_at |
AT4G21310
|
unknown |
-0.239 |
-0.131 |
| 134 |
259766_at |
AT1G64360
|
unknown |
-0.235 |
0.209 |
| 135 |
248382_at |
AT5G51890
|
[Peroxidase superfamily protein] |
-0.234 |
-0.115 |