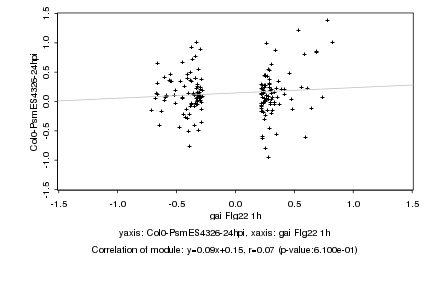
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
gai Flg22 1h |
Log2 signal ratio
Col0-PsmES4326-24hpi |
| 1 |
254234_at |
AT4G23680
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.817 |
1.018 |
| 2 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
0.781 |
1.398 |
| 3 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.736 |
0.071 |
| 4 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.688 |
0.861 |
| 5 |
245076_at |
AT2G23170
|
GH3.3 |
0.685 |
0.849 |
| 6 |
254452_at |
AT4G21100
|
DDB1B, damaged DNA binding protein 1B |
0.645 |
-0.102 |
| 7 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
0.611 |
0.226 |
| 8 |
247474_at |
AT5G62280
|
unknown |
0.592 |
-0.600 |
| 9 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
0.581 |
0.811 |
| 10 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.554 |
0.243 |
| 11 |
259866_at |
AT1G76640
|
CML39, CALMODULIN LIKE 39 |
0.532 |
1.227 |
| 12 |
245602_at |
AT4G14270
|
unknown |
0.482 |
-0.122 |
| 13 |
255811_at |
AT4G10250
|
ATHSP22.0 |
0.468 |
0.046 |
| 14 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
0.455 |
0.489 |
| 15 |
254451_at |
AT4G21090
|
ATMFDX2, ARABIDOPSIS MITOCHONDRIAL FERREDOXIN 2, MFDX2, MITOCHONDRIAL FERREDOXIN 2 |
0.415 |
0.135 |
| 16 |
245243_at |
AT1G44414
|
unknown |
0.412 |
0.206 |
| 17 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.387 |
0.217 |
| 18 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
0.369 |
-0.036 |
| 19 |
252482_at |
AT3G46670
|
UGT76E11, UDP-glucosyl transferase 76E11 |
0.364 |
0.352 |
| 20 |
262322_at |
AT1G27590
|
unknown |
0.350 |
0.069 |
| 21 |
250550_at |
AT5G07870
|
[HXXXD-type acyl-transferase family protein] |
0.348 |
0.238 |
| 22 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
0.345 |
-0.550 |
| 23 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.334 |
0.876 |
| 24 |
266392_at |
AT2G41280
|
ATM10, M10 |
0.328 |
0.170 |
| 25 |
247005_at |
AT5G67520
|
adenosine-5'-phosphosulfate (APS) kinase 4 |
0.327 |
-0.045 |
| 26 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
0.327 |
-0.013 |
| 27 |
252175_at |
AT3G50700
|
AtIDD2, indeterminate(ID)-domain 2, IDD2, indeterminate(ID)-domain 2 |
0.311 |
-0.144 |
| 28 |
252698_at |
AT3G43670
|
[Copper amine oxidase family protein] |
0.307 |
0.077 |
| 29 |
250937_at |
AT5G03230
|
unknown |
0.305 |
-0.204 |
| 30 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
0.304 |
0.638 |
| 31 |
257922_at |
AT3G23150
|
ETR2, ethylene response 2 |
0.304 |
0.141 |
| 32 |
267048_at |
AT2G34200
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.298 |
0.205 |
| 33 |
265695_at |
AT2G24490
|
ATRPA2, ATRPA32A, ROR1, SUPPRESSOR OF ROS1, RPA2, replicon protein A2, RPA32A |
0.298 |
0.039 |
| 34 |
266110_at |
AT2G02080
|
AtIDD4, indeterminate(ID)-domain 4, IDD4, indeterminate(ID)-domain 4 |
0.297 |
-0.006 |
| 35 |
250766_at |
AT5G05550
|
[sequence-specific DNA binding transcription factors] |
0.296 |
-0.048 |
| 36 |
246584_at |
AT5G14730
|
unknown |
0.290 |
0.202 |
| 37 |
264957_at |
AT1G77000
|
ATSKP2;2, ARABIDOPSIS HOMOLOG OF HOMOLOG OF HUMAN SKP2 2, SKP2B |
0.289 |
0.376 |
| 38 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
0.285 |
0.300 |
| 39 |
258897_at |
AT3G05730
|
[LOCATED IN: endomembrane system] |
0.284 |
-0.459 |
| 40 |
267337_at |
AT2G39980
|
[HXXXD-type acyl-transferase family protein] |
0.284 |
0.315 |
| 41 |
259058_at |
AT3G03470
|
CYP89A9, cytochrome P450, family 87, subfamily A, polypeptide 9 |
0.283 |
0.543 |
| 42 |
266271_at |
AT2G29440
|
ATGSTU6, glutathione S-transferase tau 6, GST24, GLUTATHIONE S-TRANSFERASE 24, GSTU6, glutathione S-transferase tau 6 |
0.281 |
0.251 |
| 43 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
0.279 |
-0.949 |
| 44 |
251971_at |
AT3G53160
|
UGT73C7, UDP-glucosyl transferase 73C7 |
0.279 |
0.556 |
| 45 |
261567_at |
AT1G33055
|
unknown |
0.277 |
0.045 |
| 46 |
245807_at |
AT1G46768
|
RAP2.1, related to AP2 1 |
0.277 |
0.100 |
| 47 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
0.268 |
-0.142 |
| 48 |
260567_at |
AT2G43820
|
ATSAGT1, Arabidopsis thaliana salicylic acid glucosyltransferase 1, GT, SAGT1, salicylic acid glucosyltransferase 1, SGT1, UDP-glucose:salicylic acid glucosyltransferase 1, UGT74F2, UDP-glucosyltransferase 74F2 |
0.265 |
0.438 |
| 49 |
266246_at |
AT2G27690
|
CYP94C1, cytochrome P450, family 94, subfamily C, polypeptide 1 |
0.261 |
1.004 |
| 50 |
266362_at |
AT2G32430
|
[Galactosyltransferase family protein] |
0.260 |
0.052 |
| 51 |
261774_at |
AT1G76260
|
DWA2, DWD (DDB1-binding WD40 protein) hypersensitive to ABA 2 |
0.258 |
0.105 |
| 52 |
247786_at |
AT5G58600
|
PMR5, POWDERY MILDEW RESISTANT 5, TBL44, TRICHOME BIREFRINGENCE-LIKE 44 |
0.255 |
0.003 |
| 53 |
247013_at |
AT5G67480
|
ATBT4, BT4, BTB and TAZ domain protein 4 |
0.255 |
0.453 |
| 54 |
264338_at |
AT1G70300
|
KUP6, K+ uptake permease 6 |
0.254 |
0.432 |
| 55 |
266300_at |
AT2G01420
|
ATPIN4, ARABIDOPSIS PIN-FORMED 4, PIN4, PIN-FORMED 4 |
0.253 |
-0.787 |
| 56 |
259877_at |
AT1G76710
|
ASHH1, ASH1 RELATED PROTEIN 1, ASHH1, ASH1-RELATED PROTEIN 1, SDG26, SET domain group 26 |
0.249 |
0.018 |
| 57 |
260365_at |
AT1G70630
|
RAY1, REDUCED ARABINOSE YARIV 1 |
0.249 |
0.275 |
| 58 |
247412_at |
AT5G63010
|
[Transducin/WD40 repeat-like superfamily protein] |
0.249 |
0.455 |
| 59 |
256225_at |
AT1G56220
|
[Dormancy/auxin associated family protein] |
0.247 |
-0.231 |
| 60 |
250363_at |
AT5G11390
|
WIT1, WPP domain-interacting protein 1 |
0.246 |
0.283 |
| 61 |
245734_at |
AT1G73480
|
[alpha/beta-Hydrolases superfamily protein] |
0.245 |
0.254 |
| 62 |
252829_at |
AT4G40060
|
ATHB-16, ATHB16, homeobox protein 16, HB16, homeobox protein 16 |
0.244 |
-0.299 |
| 63 |
258227_at |
AT3G15620
|
UVR3, UV REPAIR DEFECTIVE 3 |
0.243 |
0.063 |
| 64 |
267477_at |
AT2G02710
|
PLP, PAS/LOV PROTEIN, PLPA, PAS/LOV PROTEIN A, PLPB, PAS/LOV protein B, PLPC, PAS/LOV PROTEIN C |
0.242 |
0.453 |
| 65 |
253830_at |
AT4G27652
|
unknown |
0.238 |
0.203 |
| 66 |
249334_at |
AT5G41000
|
AtYSL4, YSL4, YELLOW STRIPE like 4 |
0.237 |
-0.176 |
| 67 |
265249_at |
AT2G01940
|
ATIDD15, ARABIDOPSIS THALIANA INDETERMINATE(ID)-DOMAIN 15, SGR5, SHOOT GRAVITROPISM 5 |
0.236 |
-0.029 |
| 68 |
247374_at |
AT5G63190
|
[MA3 domain-containing protein] |
0.234 |
0.167 |
| 69 |
250721_at |
AT5G06210
|
[RNA binding (RRM/RBD/RNP motifs) family protein] |
0.233 |
-0.016 |
| 70 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.231 |
0.139 |
| 71 |
256098_at |
AT1G13700
|
PGL1, 6-phosphogluconolactonase 1 |
0.231 |
-0.015 |
| 72 |
253030_at |
AT4G38180
|
FRS5, FAR1-related sequence 5 |
0.227 |
0.221 |
| 73 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
0.226 |
-0.624 |
| 74 |
254269_at |
AT4G23050
|
[PAS domain-containing protein tyrosine kinase family protein] |
0.226 |
0.277 |
| 75 |
249944_at |
AT5G22290
|
anac089, NAC domain containing protein 89, FSQ6, fructose-sensing quantitative trait locus 6, NAC089, NAC domain containing protein 89 |
0.225 |
0.084 |
| 76 |
265584_at |
AT2G20180
|
PIF1, PHY-INTERACTING FACTOR 1, PIL5, phytochrome interacting factor 3-like 5 |
0.225 |
-0.585 |
| 77 |
258405_at |
AT3G17590
|
BSH, BUSHY GROWTH, CHE1 |
0.223 |
-0.170 |
| 78 |
257654_at |
AT3G13310
|
DJC66, DNA J protein C66 |
0.218 |
0.298 |
| 79 |
248864_at |
AT5G46760
|
MYC3 |
0.218 |
0.231 |
| 80 |
258827_at |
AT3G07150
|
unknown |
0.217 |
0.132 |
| 81 |
250786_at |
AT5G05540
|
SDN2, small RNA degrading nuclease 2 |
0.216 |
-0.163 |
| 82 |
255943_at |
AT1G22370
|
AtUGT85A5, UDP-glucosyl transferase 85A5, UGT85A5, UDP-glucosyl transferase 85A5 |
0.216 |
-0.107 |
| 83 |
263412_at |
AT2G28720
|
[Histone superfamily protein] |
0.214 |
-0.060 |
| 84 |
255625_at |
AT4G01120
|
ATBZIP54, BASIC REGION/LEUCINE ZIPPER MOTIF 5, GBF2, G-box binding factor 2 |
0.214 |
-0.018 |
| 85 |
252387_at |
AT3G47800
|
[Galactose mutarotase-like superfamily protein] |
0.213 |
0.144 |
| 86 |
245229_at |
AT4G25620
|
[hydroxyproline-rich glycoprotein family protein] |
0.213 |
-0.083 |
| 87 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
-0.718 |
-0.140 |
| 88 |
254252_at |
AT4G23310
|
CRK23, cysteine-rich RLK (RECEPTOR-like protein kinase) 23 |
-0.682 |
0.065 |
| 89 |
253998_at |
AT4G26010
|
[Peroxidase superfamily protein] |
-0.676 |
0.147 |
| 90 |
252681_at |
AT3G44350
|
anac061, NAC domain containing protein 61, NAC061, NAC domain containing protein 61 |
-0.669 |
0.317 |
| 91 |
249940_at |
AT5G22380
|
anac090, NAC domain containing protein 90, NAC090, NAC domain containing protein 90 |
-0.667 |
0.135 |
| 92 |
266232_at |
AT2G02310
|
AtPP2-B6, phloem protein 2-B6, PP2-B6, phloem protein 2-B6 |
-0.663 |
0.662 |
| 93 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
-0.652 |
-0.392 |
| 94 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.628 |
-0.157 |
| 95 |
259850_at |
AT1G72240
|
unknown |
-0.610 |
0.424 |
| 96 |
245654_at |
AT1G56540
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.605 |
0.024 |
| 97 |
264659_at |
AT1G09930
|
ATOPT2, oligopeptide transporter 2, OPT2, oligopeptide transporter 2 |
-0.602 |
0.085 |
| 98 |
256103_at |
AT1G13540
|
unknown |
-0.593 |
0.105 |
| 99 |
256370_at |
AT1G66780
|
[MATE efflux family protein] |
-0.591 |
0.088 |
| 100 |
252596_at |
AT3G45330
|
LecRK-I.1, L-type lectin receptor kinase I.1 |
-0.563 |
0.367 |
| 101 |
266780_at |
AT2G29110
|
ATGLR2.8, glutamate receptor 2.8, GLR2.8, GLR2.8, glutamate receptor 2.8 |
-0.559 |
0.470 |
| 102 |
248775_at |
AT5G47850
|
CCR4, CRINKLY4 related 4 |
-0.556 |
0.349 |
| 103 |
249051_at |
AT5G44390
|
[FAD-binding Berberine family protein] |
-0.544 |
0.344 |
| 104 |
246616_at |
AT5G36260
|
[Eukaryotic aspartyl protease family protein] |
-0.523 |
0.116 |
| 105 |
245556_at |
AT4G15400
|
ABS1, ABNORMAL SHOOT 1, BIA1, BRASSINOSTEROID INACTIVATOR1 |
-0.520 |
0.108 |
| 106 |
245737_at |
AT1G44160
|
[HSP40/DnaJ peptide-binding protein] |
-0.511 |
-0.024 |
| 107 |
262905_at |
AT1G59730
|
ATH7, thioredoxin H-type 7, TH7, thioredoxin H-type 7 |
-0.511 |
0.192 |
| 108 |
261913_at |
AT1G65860
|
FMO GS-OX1, flavin-monooxygenase glucosinolate S-oxygenase 1 |
-0.478 |
-0.435 |
| 109 |
255342_at |
AT4G04510
|
CRK38, cysteine-rich RLK (RECEPTOR-like protein kinase) 38 |
-0.474 |
0.344 |
| 110 |
257896_at |
AT3G16920
|
ATCTL2, CTL2, chitinase-like protein 2 |
-0.455 |
0.084 |
| 111 |
247861_at |
AT5G58160
|
[actin binding] |
-0.454 |
0.068 |
| 112 |
248230_at |
AT5G53830
|
[VQ motif-containing protein] |
-0.452 |
0.669 |
| 113 |
250955_at |
AT5G03190
|
CPUORF47, conserved peptide upstream open reading frame 47 |
-0.445 |
-0.205 |
| 114 |
259325_at |
AT3G05320
|
[O-fucosyltransferase family protein] |
-0.435 |
0.263 |
| 115 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.426 |
-0.267 |
| 116 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
-0.424 |
-0.136 |
| 117 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
-0.414 |
-0.284 |
| 118 |
261476_at |
AT1G14480
|
[Ankyrin repeat family protein] |
-0.413 |
0.471 |
| 119 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
-0.413 |
0.395 |
| 120 |
266084_at |
AT2G37810
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.402 |
0.142 |
| 121 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
-0.401 |
-0.504 |
| 122 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.397 |
-0.750 |
| 123 |
255654_at |
AT4G00970
|
CRK41, cysteine-rich RLK (RECEPTOR-like protein kinase) 41 |
-0.392 |
-0.220 |
| 124 |
259418_at |
AT1G02390
|
ATGPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2, GPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2 |
-0.386 |
0.502 |
| 125 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
-0.385 |
-0.082 |
| 126 |
244901_at |
ATMG00640
|
ORF25 |
-0.384 |
0.361 |
| 127 |
249558_at |
AT5G38310
|
unknown |
-0.381 |
0.351 |
| 128 |
258183_at |
AT3G21550
|
AtDMP2, Arabidopsis thaliana DUF679 domain membrane protein 2, DMP2, DUF679 domain membrane protein 2 |
-0.380 |
-0.037 |
| 129 |
259251_at |
AT3G07600
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.377 |
0.929 |
| 130 |
260867_at |
AT1G43790
|
TED6, tracheary element differentiation-related 6 |
-0.376 |
-0.020 |
| 131 |
246373_at |
AT1G51860
|
[Leucine-rich repeat protein kinase family protein] |
-0.369 |
0.732 |
| 132 |
245300_at |
AT4G16350
|
CBL6, calcineurin B-like protein 6, SCABP2, SOS3-LIKE CALCIUM BINDING PROTEIN 2 |
-0.364 |
0.150 |
| 133 |
264200_at |
AT1G22650
|
A/N-InvD, alkaline/neutral invertase D |
-0.361 |
0.131 |
| 134 |
254773_at |
AT4G13410
|
ATCSLA15, CSLA15, CELLULOSE SYNTHASE LIKE A15 |
-0.352 |
-0.081 |
| 135 |
264078_at |
AT2G28470
|
BGAL8, beta-galactosidase 8 |
-0.349 |
-0.404 |
| 136 |
255511_at |
AT4G02075
|
PIT1, pitchoun 1 |
-0.348 |
0.106 |
| 137 |
252964_at |
AT4G38830
|
CRK26, cysteine-rich RLK (RECEPTOR-like protein kinase) 26 |
-0.345 |
0.396 |
| 138 |
258287_at |
AT3G15990
|
SULTR3;4, sulfate transporter 3;4 |
-0.343 |
0.408 |
| 139 |
249889_at |
AT5G22540
|
unknown |
-0.342 |
0.772 |
| 140 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
-0.340 |
-0.020 |
| 141 |
266309_at |
AT2G27140
|
[HSP20-like chaperones superfamily protein] |
-0.339 |
0.232 |
| 142 |
245755_at |
AT1G35210
|
unknown |
-0.339 |
1.017 |
| 143 |
267094_at |
AT2G38080
|
ATLMCO4, ARABIDOPSIS LACCASE-LIKE MULTICOPPER OXIDASE 4, IRX12, IRREGULAR XYLEM 12, LAC4, LACCASE 4, LMCO4, LACCASE-LIKE MULTICOPPER OXIDASE 4 |
-0.336 |
-0.042 |
| 144 |
263689_at |
AT1G26820
|
RNS3, ribonuclease 3 |
-0.335 |
0.058 |
| 145 |
265093_at |
AT1G03905
|
ABCI19, ATP-binding cassette I19 |
-0.330 |
0.034 |
| 146 |
262357_at |
AT1G73040
|
[Mannose-binding lectin superfamily protein] |
-0.328 |
0.292 |
| 147 |
250646_at |
AT5G06720
|
ATPA2, peroxidase 2, AtPRX53, PA2, peroxidase 2, PRX53, peroxidase 53 |
-0.327 |
0.004 |
| 148 |
253191_at |
AT4G35350
|
XCP1, xylem cysteine peptidase 1 |
-0.327 |
0.073 |
| 149 |
254402_at |
AT4G21310
|
unknown |
-0.326 |
0.170 |
| 150 |
249332_at |
AT5G40980
|
unknown |
-0.326 |
-0.040 |
| 151 |
246495_at |
AT5G16200
|
[50S ribosomal protein-related] |
-0.323 |
0.259 |
| 152 |
261016_at |
AT1G26560
|
BGLU40, beta glucosidase 40 |
-0.322 |
-0.488 |
| 153 |
267588_at |
AT2G42060
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.320 |
0.106 |
| 154 |
251297_at |
AT3G62020
|
GLP10, germin-like protein 10 |
-0.320 |
0.074 |
| 155 |
256306_at |
AT1G30370
|
DLAH, DAD1-like acylhydrolase |
-0.318 |
0.552 |
| 156 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
-0.318 |
0.148 |
| 157 |
250958_at |
AT5G03260
|
LAC11, laccase 11 |
-0.315 |
0.133 |
| 158 |
248382_at |
AT5G51890
|
[Peroxidase superfamily protein] |
-0.308 |
0.054 |
| 159 |
262231_at |
AT1G68740
|
PHO1;H1 |
-0.307 |
0.162 |
| 160 |
254905_at |
AT4G11170
|
RMG1, Resistance Methylated Gene 1 |
-0.305 |
0.902 |
| 161 |
258315_at |
AT3G16175
|
[Thioesterase superfamily protein] |
-0.302 |
0.022 |
| 162 |
247940_at |
AT5G57190
|
PSD2, phosphatidylserine decarboxylase 2 |
-0.300 |
0.213 |
| 163 |
254971_at |
AT4G10380
|
AtNIP5;1, NIP5;1, NOD26-like intrinsic protein 5;1, NLM6, NOD26-LIKE MIP 6, NLM8, NOD26-LIKE MIP 8 |
-0.300 |
0.241 |
| 164 |
260040_at |
AT1G68765
|
IDA, INFLORESCENCE DEFICIENT IN ABSCISSION |
-0.297 |
0.102 |
| 165 |
267381_at |
AT2G26190
|
[calmodulin-binding family protein] |
-0.296 |
0.069 |
| 166 |
249063_at |
AT5G44110
|
ABCI21, ATP-binding cassette A21, ATNAP2, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN, ATPOP1, POP1 |
-0.294 |
-0.016 |
| 167 |
267260_at |
AT2G23130
|
AGP17, arabinogalactan protein 17, ATAGP17 |
-0.294 |
-0.347 |
| 168 |
250153_at |
AT5G15130
|
ATWRKY72, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 72, WRKY72, WRKY DNA-binding protein 72 |
-0.290 |
0.386 |
| 169 |
263915_at |
AT2G36430
|
unknown |
-0.289 |
-0.131 |
| 170 |
245725_at |
AT1G73370
|
ATSUS6, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 6, SUS6, sucrose synthase 6 |
-0.285 |
0.090 |
| 171 |
253112_at |
AT4G35970
|
APX5, ascorbate peroxidase 5 |
-0.283 |
0.195 |