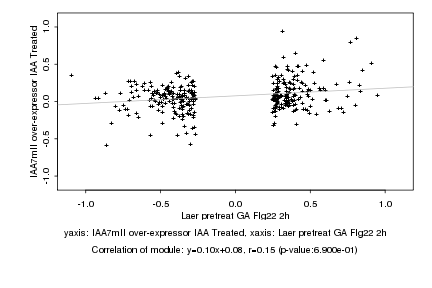
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Laer pretreat GA Flg22 2h |
Log2 signal ratio
IAA7mII over-expressor IAA Treated |
| 1 |
258957_at |
AT3G01420
|
ALPHA-DOX1, alpha-dioxygenase 1, DIOX1, DOX1, PADOX-1, plant alpha dioxygenase 1 |
0.943 |
0.086 |
| 2 |
259036_at |
AT3G09220
|
LAC7, laccase 7 |
0.903 |
0.513 |
| 3 |
251755_at |
AT3G55790
|
unknown |
0.845 |
0.420 |
| 4 |
264425_at |
AT1G61750
|
[Receptor-like protein kinase-related family protein] |
0.829 |
0.140 |
| 5 |
265342_at |
AT2G18300
|
HBI1, HOMOLOG OF BEE2 INTERACTING WITH IBH 1 |
0.823 |
0.224 |
| 6 |
260492_at |
AT2G41850
|
ADPG2, ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 2, PGAZAT, polygalacturonase abscission zone A. thaliana |
0.805 |
0.857 |
| 7 |
259316_at |
AT3G01175
|
unknown |
0.796 |
-0.047 |
| 8 |
265846_at |
AT2G35770
|
scpl28, serine carboxypeptidase-like 28 |
0.766 |
0.803 |
| 9 |
267276_at |
AT2G30130
|
ASL5, LBD12, PCK1, PEACOCK 1 |
0.758 |
0.256 |
| 10 |
253322_at |
AT4G33980
|
unknown |
0.746 |
0.071 |
| 11 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
0.716 |
-0.146 |
| 12 |
253582_at |
AT4G30670
|
[Putative membrane lipoprotein] |
0.706 |
-0.082 |
| 13 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
0.687 |
-0.089 |
| 14 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
0.670 |
0.239 |
| 15 |
251846_at |
AT3G54560
|
HTA11, histone H2A 11 |
0.628 |
-0.124 |
| 16 |
266546_at |
AT2G35270
|
AHL21, AT-hook motif nuclear localized protein 21, GIK, GIANT KILLER |
0.605 |
0.016 |
| 17 |
249154_at |
AT5G43410
|
[Integrase-type DNA-binding superfamily protein] |
0.599 |
0.153 |
| 18 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
0.595 |
0.020 |
| 19 |
266571_at |
AT2G23830
|
[PapD-like superfamily protein] |
0.584 |
0.556 |
| 20 |
265658_at |
AT2G13810
|
ALD1, AGD2-like defense response protein 1, AtALD1, EDTS5, eds two suppressor 5 |
0.582 |
0.185 |
| 21 |
246887_at |
AT5G26250
|
[Major facilitator superfamily protein] |
0.564 |
0.155 |
| 22 |
259596_at |
AT1G28130
|
GH3.17 |
0.555 |
0.176 |
| 23 |
261613_at |
AT1G49720
|
ABF1, abscisic acid responsive element-binding factor 1, AtABF1 |
0.540 |
-0.171 |
| 24 |
259442_at |
AT1G02310
|
MAN1, endo-beta-mannanase 1 |
0.525 |
0.253 |
| 25 |
264543_at |
AT1G55790
|
[FUNCTIONS IN: metal ion binding] |
0.515 |
0.401 |
| 26 |
252340_at |
AT3G48920
|
AtMYB45, myb domain protein 45, MYB45, myb domain protein 45 |
0.510 |
0.031 |
| 27 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
0.496 |
-0.061 |
| 28 |
252638_at |
AT3G44540
|
FAR4, fatty acid reductase 4 |
0.496 |
0.155 |
| 29 |
264045_at |
AT2G22450
|
AtRIBA2, RIBA2, homolog of ribA 2 |
0.491 |
-0.156 |
| 30 |
259809_at |
AT1G49800
|
unknown |
0.485 |
0.079 |
| 31 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
0.482 |
0.164 |
| 32 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.479 |
-0.111 |
| 33 |
256109_at |
AT1G16950
|
unknown |
0.478 |
0.146 |
| 34 |
260065_at |
AT1G73760
|
[RING/U-box superfamily protein] |
0.476 |
0.107 |
| 35 |
265732_at |
AT2G01300
|
unknown |
0.474 |
0.492 |
| 36 |
257154_at |
AT3G27210
|
unknown |
0.462 |
-0.106 |
| 37 |
248227_at |
AT5G53820
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.462 |
0.225 |
| 38 |
258064_at |
AT3G14680
|
CYP72A14, cytochrome P450, family 72, subfamily A, polypeptide 14 |
0.452 |
-0.094 |
| 39 |
262448_at |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
0.449 |
-0.103 |
| 40 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
0.447 |
0.253 |
| 41 |
253060_at |
AT4G37710
|
[VQ motif-containing protein] |
0.446 |
0.403 |
| 42 |
264477_at |
AT1G77240
|
AAE2, acyl activating enzyme 2 |
0.444 |
-0.024 |
| 43 |
265922_at |
AT2G18480
|
[Major facilitator superfamily protein] |
0.442 |
0.139 |
| 44 |
260887_at |
AT1G29160
|
[Dof-type zinc finger DNA-binding family protein] |
0.436 |
0.265 |
| 45 |
247710_at |
AT5G59260
|
LecRK-II.1, L-type lectin receptor kinase II.1 |
0.434 |
0.054 |
| 46 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.427 |
0.180 |
| 47 |
258760_at |
AT3G10780
|
[emp24/gp25L/p24 family/GOLD family protein] |
0.419 |
0.479 |
| 48 |
247333_at |
AT5G63600
|
ATFLS5, FLS5, flavonol synthase 5 |
0.418 |
0.039 |
| 49 |
260179_at |
AT1G70690
|
HWI1, HOPW1-1-INDUCED GENE1, PDLP5, PLASMODESMATA-LOCATED PROTEIN 5 |
0.414 |
0.359 |
| 50 |
259576_at |
AT1G35330
|
[RING/U-box superfamily protein] |
0.414 |
0.478 |
| 51 |
267572_at |
AT2G30660
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
0.413 |
0.285 |
| 52 |
256299_at |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
0.407 |
-0.296 |
| 53 |
253981_at |
AT4G26670
|
[Mitochondrial import inner membrane translocase subunit Tim17/Tim22/Tim23 family protein] |
0.407 |
-0.104 |
| 54 |
252006_at |
AT3G52820
|
ATPAP22, PURPLE ACID PHOSPHATASE 22, PAP22, purple acid phosphatase 22 |
0.399 |
0.646 |
| 55 |
267026_at |
AT2G38340
|
DREB19, dehydration response element-binding protein 19 |
0.398 |
0.082 |
| 56 |
255874_at |
AT2G40550
|
ETG1, E2F target gene 1 |
0.398 |
-0.115 |
| 57 |
249836_at |
AT5G23420
|
HMGB6, high-mobility group box 6 |
0.396 |
-0.034 |
| 58 |
263438_at |
AT2G28660
|
[Chloroplast-targeted copper chaperone protein] |
0.394 |
0.178 |
| 59 |
248744_at |
AT5G48250
|
BBX8, B-box domain protein 8 |
0.393 |
0.099 |
| 60 |
264301_at |
AT1G78780
|
[pathogenesis-related family protein] |
0.389 |
0.019 |
| 61 |
246374_at |
AT1G51840
|
[protein kinase-related] |
0.389 |
0.348 |
| 62 |
263548_at |
AT2G21660
|
ATGRP7, GLYCINE RICH PROTEIN 7, CCR2, cold, circadian rhythm, and rna binding 2, GR-RBP7, GLYCINE-RICH RNA-BINDING PROTEIN 7, GRP7, GLYCINE-RICH RNA-BINDING PROTEIN 7 |
0.387 |
-0.124 |
| 63 |
258796_at |
AT3G04630
|
WDL1, WVD2-like 1 |
0.385 |
-0.041 |
| 64 |
259975_at |
AT1G76470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.385 |
0.307 |
| 65 |
262148_at |
AT1G52560
|
[HSP20-like chaperones superfamily protein] |
0.384 |
-0.051 |
| 66 |
258338_at |
AT3G16150
|
ASPGB1, asparaginase B1 |
0.383 |
0.163 |
| 67 |
248333_at |
AT5G52390
|
[PAR1 protein] |
0.382 |
0.074 |
| 68 |
264832_at |
AT1G03660
|
[Ankyrin-repeat containing protein] |
0.382 |
0.241 |
| 69 |
250194_at |
AT5G14550
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.378 |
0.002 |
| 70 |
253743_at |
AT4G28940
|
[Phosphorylase superfamily protein] |
0.377 |
0.405 |
| 71 |
246978_at |
AT5G24910
|
CYP714A1, cytochrome P450, family 714, subfamily A, polypeptide 1, ELA1, EUI-like p450 A1 |
0.369 |
0.040 |
| 72 |
263612_at |
AT2G16440
|
MCM4, MINICHROMOSOME MAINTENANCE 4 |
0.366 |
0.247 |
| 73 |
264652_at |
AT1G08920
|
ESL1, ERD (early response to dehydration) six-like 1 |
0.364 |
0.251 |
| 74 |
245112_at |
AT2G41540
|
GPDHC1 |
0.364 |
0.167 |
| 75 |
251918_at |
AT3G54040
|
[PAR1 protein] |
0.361 |
0.268 |
| 76 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
0.361 |
-0.015 |
| 77 |
249187_at |
AT5G43060
|
RD21B, esponsive to dehydration 21B |
0.359 |
0.125 |
| 78 |
255028_at |
AT4G09890
|
unknown |
0.358 |
-0.050 |
| 79 |
261700_at |
AT1G32690
|
unknown |
0.358 |
-0.020 |
| 80 |
258813_at |
AT3G04060
|
anac046, NAC domain containing protein 46, NAC046, NAC domain containing protein 46 |
0.355 |
0.184 |
| 81 |
263681_at |
AT1G26840
|
ATORC6, ARABIDOPSIS THALIANA ORIGIN RECOGNITION COMPLEX PROTEIN 6, ORC6, origin recognition complex protein 6 |
0.353 |
0.114 |
| 82 |
249559_at |
AT5G38320
|
unknown |
0.352 |
0.054 |
| 83 |
260039_at |
AT1G68795
|
CLE12, CLAVATA3/ESR-RELATED 12 |
0.350 |
0.421 |
| 84 |
266372_at |
AT2G41310
|
ARR8, RESPONSE REGULATOR 8, ATRR3, response regulator 3, RR3, response regulator 3 |
0.350 |
-0.061 |
| 85 |
260755_at |
AT1G48980
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.349 |
0.026 |
| 86 |
259780_at |
AT1G29630
|
[5'-3' exonuclease family protein] |
0.346 |
0.436 |
| 87 |
255232_at |
AT4G05330
|
AGD13, ARF-GAP domain 13 |
0.344 |
-0.003 |
| 88 |
253871_at |
AT4G27440
|
PORB, protochlorophyllide oxidoreductase B |
0.344 |
-0.053 |
| 89 |
255607_at |
AT4G01130
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.342 |
0.470 |
| 90 |
247998_at |
AT5G56200
|
[C2H2 type zinc finger transcription factor family] |
0.341 |
0.079 |
| 91 |
255513_at |
AT4G02060
|
MCM7, PRL, PROLIFERA |
0.339 |
0.090 |
| 92 |
247028_at |
AT5G67100
|
ICU2, INCURVATA2 |
0.339 |
0.105 |
| 93 |
248410_at |
AT5G51570
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
0.338 |
-0.034 |
| 94 |
248685_at |
AT5G48500
|
unknown |
0.338 |
0.041 |
| 95 |
251114_at |
AT5G01380
|
[Homeodomain-like superfamily protein] |
0.336 |
0.145 |
| 96 |
255252_at |
AT4G04990
|
unknown |
0.335 |
0.191 |
| 97 |
252527_at |
AT3G46440
|
UXS5, UDP-XYL synthase 5 |
0.335 |
0.007 |
| 98 |
248139_at |
AT5G54970
|
unknown |
0.333 |
0.236 |
| 99 |
261883_at |
AT1G80870
|
[Protein kinase superfamily protein] |
0.332 |
-0.022 |
| 100 |
258953_at |
AT3G01430
|
unknown |
0.332 |
-0.047 |
| 101 |
256715_at |
AT2G34090
|
MEE18, maternal effect embryo arrest 18 |
0.332 |
0.075 |
| 102 |
249580_at |
AT5G37740
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.329 |
-0.021 |
| 103 |
249882_at |
AT5G22890
|
STOP2, sensitive to proton rhizotoxicity 2 |
0.327 |
0.023 |
| 104 |
256617_at |
AT3G22240
|
unknown |
0.324 |
0.264 |
| 105 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.320 |
0.594 |
| 106 |
256833_at |
AT3G22910
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.319 |
0.301 |
| 107 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
0.318 |
-0.087 |
| 108 |
264535_at |
AT1G55690
|
[Sec14p-like phosphatidylinositol transfer family protein] |
0.318 |
0.244 |
| 109 |
266232_at |
AT2G02310
|
AtPP2-B6, phloem protein 2-B6, PP2-B6, phloem protein 2-B6 |
0.311 |
0.944 |
| 110 |
259282_at |
AT3G11430
|
ATGPAT5, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 5, GPAT5, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 5 |
0.306 |
0.362 |
| 111 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
0.304 |
-0.078 |
| 112 |
257367_at |
AT2G25780
|
unknown |
0.302 |
0.072 |
| 113 |
245064_at |
AT2G39725
|
[LYR family of Fe/S cluster biogenesis protein] |
0.302 |
-0.035 |
| 114 |
257946_at |
AT3G21710
|
unknown |
0.302 |
0.082 |
| 115 |
266271_at |
AT2G29440
|
ATGSTU6, glutathione S-transferase tau 6, GST24, GLUTATHIONE S-TRANSFERASE 24, GSTU6, glutathione S-transferase tau 6 |
0.296 |
0.092 |
| 116 |
249009_at |
AT5G44610
|
MAP18, microtubule-associated protein 18, PCAP2, PLASMA MEMBRANE ASSOCIATED CA2+-BINDING PROTEIN-2 |
0.295 |
0.082 |
| 117 |
257024_at |
AT3G19100
|
[Protein kinase superfamily protein] |
0.294 |
-0.078 |
| 118 |
258005_at |
AT3G19390
|
[Granulin repeat cysteine protease family protein] |
0.293 |
0.056 |
| 119 |
249839_at |
AT5G23405
|
[HMG-box (high mobility group) DNA-binding family protein] |
0.291 |
-0.096 |
| 120 |
259073_at |
AT3G02290
|
[RING/U-box superfamily protein] |
0.289 |
0.279 |
| 121 |
249362_at |
AT5G40550
|
AtSGF29b, SGF29b, SaGa associated Factor 29 b |
0.287 |
0.325 |
| 122 |
266352_at |
AT2G01610
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.287 |
0.053 |
| 123 |
264978_at |
AT1G27120
|
[Galactosyltransferase family protein] |
0.282 |
0.181 |
| 124 |
265116_at |
AT1G62480
|
[Vacuolar calcium-binding protein-related] |
0.282 |
0.080 |
| 125 |
252653_at |
AT3G44730
|
AtKIN14h, ATKP1, ARABIDOPSIS KINESIN-LIKE PROTEIN 1, KP1, kinesin-like protein 1 |
0.281 |
0.142 |
| 126 |
254803_at |
AT4G13100
|
[RING/U-box superfamily protein] |
0.281 |
0.188 |
| 127 |
250860_at |
AT5G04770
|
ATCAT6, ARABIDOPSIS THALIANA CATIONIC AMINO ACID TRANSPORTER 6, CAT6, cationic amino acid transporter 6 |
0.280 |
-0.021 |
| 128 |
249916_at |
AT5G22880
|
H2B, HISTONE H2B, HTB2, histone B2 |
0.280 |
0.085 |
| 129 |
247525_at |
AT5G61380
|
APRR1, AtTOC1, TIMING OF CAB EXPRESSION 1, PRR1, PSEUDO-RESPONSE REGULATOR 1, TOC1, TIMING OF CAB EXPRESSION 1 |
0.279 |
-0.007 |
| 130 |
260674_at |
AT1G19370
|
unknown |
0.279 |
0.012 |
| 131 |
249087_at |
AT5G44210
|
ATERF-9, ERF DOMAIN PROTEIN- 9, ATERF9, ERF DOMAIN PROTEIN 9, ERF9, erf domain protein 9 |
0.278 |
0.254 |
| 132 |
248891_at |
AT5G46280
|
MCM3, MINICHROMOSOME MAINTENANCE 3 |
0.278 |
0.202 |
| 133 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
0.276 |
0.127 |
| 134 |
257541_at |
AT3G25950
|
[TRAM, LAG1 and CLN8 (TLC) lipid-sensing domain containing protein] |
0.276 |
0.300 |
| 135 |
264211_at |
AT1G22770
|
FB, GI, GIGANTEA |
0.275 |
0.042 |
| 136 |
266839_at |
AT2G25930
|
ELF3, EARLY FLOWERING 3, PYK20 |
0.274 |
0.037 |
| 137 |
266938_at |
AT2G18950
|
ATHPT, HOMOGENTISATE PHYTYLTRANSFERASE, HPT1, homogentisate phytyltransferase 1, TPT1, VTE2, VITAMIN E 2 |
0.274 |
0.459 |
| 138 |
260728_at |
AT1G48210
|
[Protein kinase superfamily protein] |
0.273 |
0.025 |
| 139 |
265323_at |
AT2G18260
|
ATSYP112, SYP112, syntaxin of plants 112 |
0.272 |
0.087 |
| 140 |
264638_at |
AT1G65480
|
FT, FLOWERING LOCUS T, RSB8, REDUCED STEM BRANCHING 8 |
0.272 |
0.020 |
| 141 |
250024_at |
AT5G18270
|
ANAC087, Arabidopsis NAC domain containing protein 87 |
0.271 |
0.186 |
| 142 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
0.271 |
-0.056 |
| 143 |
246080_at |
AT5G20460
|
unknown |
0.268 |
0.173 |
| 144 |
262881_at |
AT1G64890
|
[Major facilitator superfamily protein] |
0.268 |
-0.030 |
| 145 |
251494_at |
AT3G59350
|
[Protein kinase superfamily protein] |
0.267 |
-0.050 |
| 146 |
262296_at |
AT1G27630
|
CYCT1;3, cyclin T 1;3 |
0.266 |
0.020 |
| 147 |
256430_at |
AT3G11020
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2B, DRE/CRT-binding protein 2B |
0.265 |
0.014 |
| 148 |
267178_at |
AT2G37750
|
unknown |
0.265 |
0.109 |
| 149 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
0.265 |
0.105 |
| 150 |
245644_at |
AT1G25320
|
[Leucine-rich repeat protein kinase family protein] |
0.265 |
-0.090 |
| 151 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
0.265 |
-0.068 |
| 152 |
262244_at |
AT1G48260
|
CIPK17, CBL-interacting protein kinase 17, SnRK3.21, SNF1-RELATED PROTEIN KINASE 3.21 |
0.264 |
0.359 |
| 153 |
245393_at |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
0.262 |
0.478 |
| 154 |
260412_at |
AT1G69830
|
AMY3, alpha-amylase-like 3, ATAMY3, ALPHA-AMYLASE-LIKE 3 |
0.261 |
-0.133 |
| 155 |
267254_at |
AT2G23030
|
SNRK2-9, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-9, SNRK2.9, SNF1-related protein kinase 2.9 |
0.261 |
-0.189 |
| 156 |
263412_at |
AT2G28720
|
[Histone superfamily protein] |
0.261 |
0.087 |
| 157 |
267355_at |
AT2G39900
|
WLIM2a, WLIM2a |
0.260 |
-0.133 |
| 158 |
265321_at |
AT2G18280
|
AtTLP2, tubby like protein 2, TLP2, tubby like protein 2 |
0.259 |
0.026 |
| 159 |
251132_at |
AT5G01200
|
[Duplicated homeodomain-like superfamily protein] |
0.259 |
-0.284 |
| 160 |
247867_at |
AT5G57630
|
CIPK21, CBL-interacting protein kinase 21, SnRK3.4, SNF1-RELATED PROTEIN KINASE 3.4 |
0.259 |
0.050 |
| 161 |
253243_at |
AT4G34560
|
unknown |
0.259 |
0.072 |
| 162 |
261272_at |
AT1G26665
|
[Mediator complex, subunit Med10] |
0.258 |
0.059 |
| 163 |
251421_at |
AT3G60510
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
0.256 |
0.087 |
| 164 |
260636_at |
AT1G62430
|
ATCDS1, CDP-diacylglycerol synthase 1, CDS1, CDP-diacylglycerol synthase 1 |
0.254 |
0.084 |
| 165 |
261838_at |
AT1G16030
|
Hsp70b, heat shock protein 70B |
0.254 |
-0.092 |
| 166 |
260171_at |
AT1G71910
|
unknown |
0.254 |
-0.312 |
| 167 |
246155_at |
AT5G20030
|
[Plant Tudor-like RNA-binding protein] |
0.252 |
0.019 |
| 168 |
267378_at |
AT2G26200
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.251 |
0.046 |
| 169 |
247045_at |
AT5G66930
|
unknown |
0.250 |
0.153 |
| 170 |
249800_at |
AT5G23660
|
AtSWEET12, MTN3, homolog of Medicago truncatula MTN3, SWEET12 |
0.250 |
0.243 |
| 171 |
246288_at |
AT1G31850
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.250 |
-0.314 |
| 172 |
250935_at |
AT5G03240
|
UBQ3, polyubiquitin 3 |
0.250 |
0.015 |
| 173 |
252076_at |
AT3G51660
|
[Tautomerase/MIF superfamily protein] |
0.247 |
0.349 |
| 174 |
256282_at |
AT3G12550
|
FDM3, factor of DNA methylation 3 |
0.247 |
-0.145 |
| 175 |
247444_at |
AT5G62630
|
HIPL2, hipl2 protein precursor |
0.247 |
0.037 |
| 176 |
266421_at |
AT2G38540
|
ATLTP1, ARABIDOPSIS THALIANA LIPID TRANSFER PROTEIN 1, LP1, lipid transfer protein 1, LTP1, LIPID TRANSFER PROTEIN 1 |
-1.100 |
0.355 |
| 177 |
256598_at |
AT3G30180
|
BR6OX2, brassinosteroid-6-oxidase 2, CYP85A2 |
-0.939 |
0.047 |
| 178 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
-0.918 |
0.051 |
| 179 |
260267_at |
AT1G68530
|
CER6, ECERIFERUM 6, CUT1, CUTICULAR 1, G2, KCS6, 3-ketoacyl-CoA synthase 6, POP1, POLLEN-PISTIL INCOMPATIBILITY 1 |
-0.874 |
0.108 |
| 180 |
259592_at |
AT1G27950
|
LTPG1, glycosylphosphatidylinositol-anchored lipid protein transfer 1 |
-0.864 |
-0.585 |
| 181 |
259892_at |
AT1G72610
|
ATGER1, A. THALIANA GERMIN-LIKE PROTEIN 1, GER1, germin-like protein 1, GLP1, GERMIN-LIKE PROTEIN 1 |
-0.830 |
-0.287 |
| 182 |
263431_at |
AT2G22170
|
PLAT2, PLAT domain protein 2 |
-0.805 |
-0.063 |
| 183 |
263664_at |
AT1G04250
|
AXR3, AUXIN RESISTANT 3, IAA17, indole-3-acetic acid inducible 17 |
-0.779 |
-0.118 |
| 184 |
255690_at |
AT4G00360
|
ATT1, ABERRANT INDUCTION OF TYPE THREE 1, CYP86A2, cytochrome P450, family 86, subfamily A, polypeptide 2 |
-0.769 |
0.114 |
| 185 |
256880_at |
AT3G26450
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.752 |
-0.047 |
| 186 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
-0.740 |
-0.096 |
| 187 |
256870_at |
AT3G26300
|
CYP71B34, cytochrome P450, family 71, subfamily B, polypeptide 34 |
-0.722 |
-0.106 |
| 188 |
257938_at |
AT3G19820
|
CBB1, CABBAGE 1, DIM, DIMINUTIA, DIM1, DIMINUTO 1, DWF1, DWARF 1, EVE1, ENHANCED VERY-LOW-FLUENCE RESPONSES 1 |
-0.717 |
-0.181 |
| 189 |
254785_at |
AT4G12730
|
FLA2, FASCICLIN-like arabinogalactan 2 |
-0.717 |
0.271 |
| 190 |
262288_at |
AT1G70760
|
CRR23, CHLORORESPIRATORY REDUCTION 23, NdhL, NADH dehydrogenase-like complex L |
-0.712 |
0.014 |
| 191 |
260140_at |
AT1G66390
|
ATMYB90, MYB90, myb domain protein 90, PAP2, PRODUCTION OF ANTHOCYANIN PIGMENT 2 |
-0.703 |
0.273 |
| 192 |
255786_at |
AT1G19670
|
ATCLH1, chlorophyllase 1, ATHCOR1, CORONATINE-INDUCED PROTEIN 1, CLH1, chlorophyllase 1, CORI1, CORONATINE-INDUCED PROTEIN 1 |
-0.700 |
0.127 |
| 193 |
252366_at |
AT3G48420
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.698 |
0.209 |
| 194 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
-0.686 |
0.060 |
| 195 |
252115_at |
AT3G51600
|
LTP5, lipid transfer protein 5 |
-0.675 |
0.277 |
| 196 |
256825_at |
AT3G22120
|
CWLP, cell wall-plasma membrane linker protein |
-0.667 |
-0.149 |
| 197 |
253372_at |
AT4G33220
|
ATPME44, A. THALIANA PECTIN METHYLESTERASE 44, PME44, pectin methylesterase 44 |
-0.664 |
0.148 |
| 198 |
259430_at |
AT1G01610
|
ATGPAT4, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 4, GPAT4, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 4 |
-0.661 |
0.229 |
| 199 |
262903_at |
AT1G59950
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.654 |
0.080 |
| 200 |
263709_at |
AT1G09310
|
unknown |
-0.648 |
-0.202 |
| 201 |
255889_at |
AT1G17840
|
ABCG11, ATP-binding cassette G11, AtABCG11, ATWBC11, ARABIDOPSIS THALIANA WHITE-BROWN COMPLEX HOMOLOG PROTEIN 11, COF1, CUTICULAR DEFECT AND ORGAN FUSION 1, DSO, DESPERADO, WBC11, white-brown complex homolog protein 11 |
-0.626 |
0.208 |
| 202 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
-0.614 |
0.149 |
| 203 |
246701_at |
AT5G28020
|
ATCYSD2, CYSTEINE SYNTHASE D2, CYSD2, cysteine synthase D2 |
-0.611 |
0.242 |
| 204 |
262281_at |
AT1G68570
|
AtNPF3.1, NPF3.1, NRT1/ PTR family 3.1 |
-0.587 |
0.157 |
| 205 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.577 |
0.022 |
| 206 |
254034_at |
AT4G25960
|
ABCB2, ATP-binding cassette B2, PGP2, P-glycoprotein 2 |
-0.574 |
0.257 |
| 207 |
258033_at |
AT3G21250
|
ABCC8, ATP-binding cassette C8, ATMRP6, ARABIDOPSIS THALIANA MULTIDRUG RESISTANCE-ASSOCIATED PROTEIN 6, MRP6, multidrug resistance-associated protein 6 |
-0.569 |
-0.452 |
| 208 |
262577_at |
AT1G15290
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.568 |
-0.058 |
| 209 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
-0.565 |
0.052 |
| 210 |
248153_at |
AT5G54250
|
ATCNGC4, cyclic nucleotide-gated cation channel 4, CNGC4, cyclic nucleotide-gated cation channel 4, DND2, DEFENSE, NO DEATH 2, HLM1 |
-0.564 |
0.202 |
| 211 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
-0.558 |
0.120 |
| 212 |
254535_at |
AT4G19710
|
AK-HSDH, ASPARTATE KINASE-HOMOSERINE DEHYDROGENASE, AK-HSDH II, aspartate kinase-homoserine dehydrogenase ii |
-0.557 |
-0.058 |
| 213 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
-0.555 |
0.120 |
| 214 |
252604_at |
AT3G45060
|
ATNRT2.6, ARABIDOPSIS THALIANA HIGH AFFINITY NITRATE TRANSPORTER 2.6, NRT2.6, high affinity nitrate transporter 2.6 |
-0.549 |
0.083 |
| 215 |
252412_at |
AT3G47295
|
unknown |
-0.543 |
0.014 |
| 216 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
-0.542 |
0.144 |
| 217 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-0.540 |
0.007 |
| 218 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
-0.529 |
0.115 |
| 219 |
255298_at |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
-0.523 |
0.125 |
| 220 |
250261_at |
AT5G13400
|
[Major facilitator superfamily protein] |
-0.522 |
0.158 |
| 221 |
259608_at |
AT1G27960
|
ECT9, evolutionarily conserved C-terminal region 9 |
-0.520 |
-0.119 |
| 222 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
-0.514 |
-0.033 |
| 223 |
259579_at |
AT1G28010
|
ABCB14, ATP-binding cassette B14, ATABCB14, Arabidopsis thaliana ATP-binding cassette B14, MDR12, multi-drug resistance 12, PGP14, P-glycoprotein 14 |
-0.508 |
0.100 |
| 224 |
246339_at |
AT3G44890
|
RPL9, ribosomal protein L9 |
-0.508 |
-0.002 |
| 225 |
247563_at |
AT5G61130
|
AtPDCB1, PDCB1, plasmodesmata callose-binding protein 1 |
-0.502 |
0.059 |
| 226 |
247218_at |
AT5G65010
|
ASN2, asparagine synthetase 2 |
-0.498 |
-0.010 |
| 227 |
245612_at |
AT4G14440
|
ATECI3, ARABIDOPSIS THALIANA DELTA(3), DELTA(2)-ENOYL COA ISOMERASE 3, ECI3, DELTA(3), DELTA(2)-ENOYL COA ISOMERASE 3, HCD1, 3-hydroxyacyl-CoA dehydratase 1 |
-0.493 |
0.228 |
| 228 |
267377_at |
AT2G26250
|
FDH, FIDDLEHEAD, KCS10, 3-ketoacyl-CoA synthase 10 |
-0.489 |
0.090 |
| 229 |
251317_at |
AT3G61490
|
[Pectin lyase-like superfamily protein] |
-0.489 |
-0.059 |
| 230 |
258764_at |
AT3G10720
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.488 |
-0.291 |
| 231 |
266415_at |
AT2G38530
|
cdf3, cell growth defect factor-3, LP2, LTP2, lipid transfer protein 2 |
-0.488 |
-0.142 |
| 232 |
261881_at |
AT1G80760
|
NIP6, NIP6;1, NOD26-like intrinsic protein 6;1, NLM7 |
-0.482 |
0.034 |
| 233 |
254041_at |
AT4G25830
|
[Uncharacterised protein family (UPF0497)] |
-0.480 |
0.119 |
| 234 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
-0.475 |
0.097 |
| 235 |
248619_at |
AT5G49630
|
AAP6, amino acid permease 6 |
-0.473 |
0.070 |
| 236 |
266856_at |
AT2G26910
|
ABCG32, ATP-binding cassette G32, AtABCG32, ATPDR4, PLEIOTROPIC DRUG RESISTANCE 4, PDR4, pleiotropic drug resistance 4, PEC1, PERMEABLE CUTICLE 1 |
-0.472 |
0.172 |
| 237 |
265823_at |
AT2G35760
|
[Uncharacterised protein family (UPF0497)] |
-0.472 |
-0.014 |
| 238 |
245029_at |
AT2G26580
|
YAB5, YABBY5 |
-0.465 |
0.042 |
| 239 |
260840_at |
AT1G29050
|
TBL38, TRICHOME BIREFRINGENCE-LIKE 38 |
-0.458 |
0.198 |
| 240 |
266215_at |
AT2G06850
|
EXGT-A1, endoxyloglucan transferase A1, EXT, ENDOXYLOGLUCAN TRANSFERASE, XTH4, xyloglucan endotransglucosylase/hydrolase 4 |
-0.455 |
0.178 |
| 241 |
252487_at |
AT3G46660
|
UGT76E12, UDP-glucosyl transferase 76E12 |
-0.452 |
0.202 |
| 242 |
261221_at |
AT1G19960
|
unknown |
-0.451 |
0.140 |
| 243 |
260490_at |
AT1G51500
|
ABCG12, ATP-binding cassette G12, AtABCG12, ATWBC12, ARABIDOPSIS THALIANA WHITE-BROWN COMPLEX 12, CER5, ECERIFERUM 5, D3, WBC12, WHITE-BROWN COMPLEX 12 |
-0.450 |
0.153 |
| 244 |
261826_at |
AT1G11580
|
ATPMEPCRA, PMEPCRA, methylesterase PCR A |
-0.445 |
0.114 |
| 245 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
-0.442 |
0.107 |
| 246 |
255732_at |
AT1G25450
|
CER60, ECERIFERUM 60, KCS5, 3-ketoacyl-CoA synthase 5 |
-0.442 |
0.106 |
| 247 |
261981_at |
AT1G33811
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.441 |
0.011 |
| 248 |
261375_at |
AT1G53160
|
FTM6, FLORAL TRANSITION AT THE MERISTEM6, SPL4, squamosa promoter binding protein-like 4 |
-0.434 |
0.131 |
| 249 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
-0.432 |
0.147 |
| 250 |
267472_at |
AT2G02850
|
ARPN, plantacyanin |
-0.431 |
0.262 |
| 251 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
-0.427 |
-0.016 |
| 252 |
247529_at |
AT5G61520
|
[Major facilitator superfamily protein] |
-0.426 |
0.077 |
| 253 |
265042_at |
AT1G04040
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.422 |
0.032 |
| 254 |
259383_at |
AT3G16470
|
JAL35, jacalin-related lectin 35, JR1, JASMONATE RESPONSIVE 1 |
-0.420 |
-0.075 |
| 255 |
251353_at |
AT3G61080
|
[Protein kinase superfamily protein] |
-0.417 |
0.114 |
| 256 |
262563_at |
AT1G34210
|
ATSERK2, SERK2, somatic embryogenesis receptor-like kinase 2 |
-0.417 |
-0.220 |
| 257 |
265682_at |
AT2G24390
|
[AIG2-like (avirulence induced gene) family protein] |
-0.416 |
0.037 |
| 258 |
246036_at |
AT5G08370
|
AGAL2, alpha-galactosidase 2, AtAGAL2, alpha-galactosidase 2 |
-0.416 |
0.036 |
| 259 |
259839_at |
AT1G52190
|
AtNPF1.2, NPF1.2, NRT1/ PTR family 1.2, NRT1.11, nitrate transporter 1.11 |
-0.415 |
0.197 |
| 260 |
252863_at |
AT4G39800
|
ATIPS1, INOSITOL 3-PHOSPHATE SYNTHASE 1, ATMIPS1, MYO-INOSITOL-1-PHOSPHATE SYNTHASE 1, MI-1-P SYNTHASE, MIPS1, D-myo-Inositol 3-Phosphate Synthase 1, MIPS1, myo-inositol-1-phosphate synthase 1 |
-0.409 |
-0.026 |
| 261 |
262582_at |
AT1G15410
|
[aspartate-glutamate racemase family] |
-0.409 |
-0.127 |
| 262 |
256304_at |
AT1G69523
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.405 |
-0.158 |
| 263 |
246097_at |
AT5G20270
|
HHP1, heptahelical transmembrane protein1 |
-0.398 |
0.389 |
| 264 |
256076_at |
AT1G18060
|
unknown |
-0.395 |
0.049 |
| 265 |
264357_at |
AT1G03360
|
ATRRP4, ribosomal RNA processing 4, RRP4, ribosomal RNA processing 4 |
-0.394 |
-0.162 |
| 266 |
249134_at |
AT5G43150
|
unknown |
-0.392 |
-0.450 |
| 267 |
255957_at |
AT1G22160
|
unknown |
-0.392 |
0.048 |
| 268 |
251154_at |
AT3G63110
|
ATIPT3, isopentenyltransferase 3, IPT3, isopentenyltransferase 3 |
-0.388 |
0.098 |
| 269 |
245701_at |
AT5G04140
|
FD-GOGAT, FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE, GLS1, FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE 1, GLU1, glutamate synthase 1, GLUS |
-0.386 |
0.395 |
| 270 |
245816_at |
AT1G26210
|
ATSOFL1, SOB FIVE-LIKE 1, SOFL1, SOB five-like 1 |
-0.378 |
-0.091 |
| 271 |
262736_at |
AT1G28570
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.378 |
0.106 |
| 272 |
257547_at |
AT3G13000
|
unknown |
-0.377 |
0.041 |
| 273 |
263696_at |
AT1G31230
|
AK-HSDH, ASPARTATE KINASE-HOMOSERINE DEHYDROGENASE, AK-HSDH I, aspartate kinase-homoserine dehydrogenase i |
-0.377 |
-0.197 |
| 274 |
252711_at |
AT3G43720
|
LTPG2, glycosylphosphatidylinositol-anchored lipid protein transfer 2 |
-0.376 |
0.336 |
| 275 |
263809_at |
AT2G04570
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.375 |
0.068 |
| 276 |
248186_at |
AT5G53880
|
unknown |
-0.374 |
0.062 |
| 277 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.373 |
0.159 |
| 278 |
249011_at |
AT5G44670
|
GALS2, galactan synthase 2 |
-0.372 |
0.157 |
| 279 |
261224_at |
AT1G20160
|
ATSBT5.2 |
-0.372 |
-0.013 |
| 280 |
264200_at |
AT1G22650
|
A/N-InvD, alkaline/neutral invertase D |
-0.368 |
0.196 |
| 281 |
267414_at |
AT2G34790
|
EDA28, EMBRYO SAC DEVELOPMENT ARREST 28, MEE23, MATERNAL EFFECT EMBRYO ARREST 23 |
-0.366 |
0.002 |
| 282 |
254242_at |
AT4G23200
|
CRK12, cysteine-rich RLK (RECEPTOR-like protein kinase) 12 |
-0.366 |
0.034 |
| 283 |
263209_at |
AT1G10522
|
PRIN2, PLASTID REDOX INSENSITIVE 2 |
-0.363 |
-0.040 |
| 284 |
262870_at |
AT1G64710
|
[GroES-like zinc-binding dehydrogenase family protein] |
-0.361 |
0.211 |
| 285 |
250936_at |
AT5G03120
|
unknown |
-0.360 |
0.091 |
| 286 |
262313_at |
AT1G70900
|
unknown |
-0.359 |
-0.080 |
| 287 |
254122_at |
AT4G24510
|
CER2, ECERIFERUM 2, VC-2, VC2 |
-0.357 |
-0.097 |
| 288 |
263491_at |
AT2G42600
|
ATPPC2, phosphoenolpyruvate carboxylase 2, PPC2, phosphoenolpyruvate carboxylase 2 |
-0.355 |
-0.167 |
| 289 |
250243_at |
AT5G13630
|
ABAR, ABA-BINDING PROTEIN, CCH, CONDITIONAL CHLORINA, CCH1, CHLH, H SUBUNIT OF MG-CHELATASE, GUN5, GENOMES UNCOUPLED 5 |
-0.355 |
-0.232 |
| 290 |
253911_at |
AT4G27300
|
[S-locus lectin protein kinase family protein] |
-0.354 |
0.070 |
| 291 |
264313_at |
AT1G70410
|
ATBCA4, BETA CARBONIC ANHYDRASE 4, BCA4, beta carbonic anhydrase 4, CA4, BETA CARBONIC ANHYDRASE 4 |
-0.354 |
0.039 |
| 292 |
258920_at |
AT3G10520
|
AHB2, haemoglobin 2, ARATH GLB2, ATGLB2, ARABIDOPSIS HEMOGLOBIN 2, GLB2, HEMOGLOBIN 2, HB2, haemoglobin 2, NSHB2, NON-SYMBIOTIC HAEMOGLOBIN 2 |
-0.352 |
-0.057 |
| 293 |
247101_at |
AT5G66530
|
[Galactose mutarotase-like superfamily protein] |
-0.348 |
-0.197 |
| 294 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
-0.347 |
0.010 |
| 295 |
255506_at |
AT4G02130
|
GATL6, galacturonosyltransferase-like 6, LGT10 |
-0.346 |
0.093 |
| 296 |
254126_at |
AT4G24770
|
ATRBP31, ARABIDOPSIS THALIANA RNA BINDING PROTEIN, APPROXIMATELY 31 KD, ATRBP33, CP31, RBP31, 31-kDa RNA binding protein |
-0.346 |
0.111 |
| 297 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
-0.345 |
0.084 |
| 298 |
250495_at |
AT5G09770
|
[Ribosomal protein L17 family protein] |
-0.345 |
0.116 |
| 299 |
263477_at |
AT2G31790
|
[UDP-Glycosyltransferase superfamily protein] |
-0.345 |
0.153 |
| 300 |
262913_at |
AT1G59960
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.343 |
-0.333 |
| 301 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
-0.339 |
0.310 |
| 302 |
263678_at |
AT1G04420
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.331 |
0.064 |
| 303 |
263947_at |
AT2G35820
|
[ureidoglycolate hydrolases] |
-0.328 |
0.047 |
| 304 |
246076_at |
AT5G20280
|
ATSPS1F, sucrose phosphate synthase 1F, SPS1F, sucrose phosphate synthase 1F, SPSA1, sucrose-phosphate synthase A1 |
-0.328 |
-0.032 |
| 305 |
252184_at |
AT3G50660
|
AtDWF4, CLM, CLOMAZONE-RESISTANT, CYP90B1, CYTOCHROME P450 90B1, DWF4, DWARF 4, PSC1, PARTIALLY SUPPRESSING COI1 INSENSITIVITY TO JA 1, SAV1, SHADE AVOIDANCE 1, SNP2, SUPPRESSOR OF NPH4 2 |
-0.327 |
-0.420 |
| 306 |
263760_at |
AT2G21280
|
ATSULA, GC1, GIANT CHLOROPLAST 1, SULA |
-0.326 |
-0.034 |
| 307 |
258263_at |
AT3G15780
|
unknown |
-0.323 |
-0.152 |
| 308 |
262247_at |
AT1G48420
|
ACD1, 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID DEAMINASE 1, ATACD1, A. THALIANA 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID DEAMINASE 1, AtDCD, D-CDES, D-cysteine desulfhydrase, DCD, D-cysteine desulfhydrase |
-0.321 |
-0.158 |
| 309 |
256222_at |
AT1G56210
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.318 |
0.051 |
| 310 |
259707_at |
AT1G77490
|
TAPX, thylakoidal ascorbate peroxidase |
-0.318 |
0.226 |
| 311 |
255104_at |
AT4G08685
|
SAH7 |
-0.317 |
0.136 |
| 312 |
265831_at |
AT2G14460
|
unknown |
-0.315 |
-0.038 |
| 313 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
-0.315 |
0.349 |
| 314 |
245362_at |
AT4G17460
|
HAT1, JAB, JAIBA |
-0.314 |
-0.161 |
| 315 |
262703_at |
AT1G16510
|
SAUR41, SMALL AUXIN UPREGULATED 41 |
-0.312 |
-0.110 |
| 316 |
254798_at |
AT4G13050
|
[Acyl-ACP thioesterase] |
-0.309 |
-0.044 |
| 317 |
254667_at |
AT4G18280
|
[glycine-rich cell wall protein-related] |
-0.308 |
0.001 |
| 318 |
250446_at |
AT5G10770
|
[Eukaryotic aspartyl protease family protein] |
-0.307 |
0.159 |
| 319 |
253749_at |
AT4G29080
|
IAA27, indole-3-acetic acid inducible 27, PAP2, phytochrome-associated protein 2 |
-0.305 |
-0.174 |
| 320 |
257911_at |
AT3G25530
|
ATGHBDH, AtGLYR1, GHBDH, GLYR1, glyoxylate reductase 1, GR1, GLYOXYLATE REDUCTASE 1 |
-0.302 |
-0.570 |
| 321 |
263410_at |
AT2G04039
|
unknown |
-0.299 |
0.124 |
| 322 |
264466_at |
AT1G10380
|
[Putative membrane lipoprotein] |
-0.297 |
0.259 |
| 323 |
254402_at |
AT4G21310
|
unknown |
-0.297 |
0.161 |
| 324 |
245008_at |
ATCG00360
|
YCF3 |
-0.295 |
0.036 |
| 325 |
251858_at |
AT3G54820
|
PIP2;5, plasma membrane intrinsic protein 2;5, PIP2D, PLASMA MEMBRANE INTRINSIC PROTEIN 2D |
-0.294 |
0.111 |
| 326 |
256983_at |
AT3G13470
|
Cpn60beta2, chaperonin-60beta2 |
-0.294 |
-0.092 |
| 327 |
252943_at |
AT4G39330
|
ATCAD9, CAD9, cinnamyl alcohol dehydrogenase 9 |
-0.292 |
0.061 |
| 328 |
254153_at |
AT4G24450
|
ATGWD2, GWD3, PWD, phosphoglucan, water dikinase |
-0.291 |
0.044 |
| 329 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
-0.290 |
0.282 |
| 330 |
250610_at |
AT5G07550
|
ATGRP19, GRP19, glycine-rich protein 19 |
-0.290 |
0.124 |
| 331 |
264007_at |
AT2G21140
|
ATPRP2, proline-rich protein 2, PRP2, proline-rich protein 2 |
-0.288 |
0.226 |
| 332 |
246454_at |
AT5G16710
|
DHAR3, dehydroascorbate reductase 1 |
-0.287 |
-0.353 |
| 333 |
264527_at |
AT1G30760
|
[FAD-binding Berberine family protein] |
-0.286 |
-0.038 |
| 334 |
258457_at |
AT3G22425
|
HISN5A, IGPD, imidazoleglycerol-phosphate dehydratase |
-0.285 |
0.029 |
| 335 |
253131_at |
AT4G35550
|
ATWOX13, HB-4, WOX13, WUSCHEL related homeobox 13 |
-0.285 |
-0.175 |
| 336 |
256255_at |
AT3G11280
|
[Duplicated homeodomain-like superfamily protein] |
-0.282 |
0.273 |
| 337 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
-0.281 |
0.089 |
| 338 |
257071_at |
AT3G28180
|
ATCSLC04, Cellulose-synthase-like C4, ATCSLC4, CSLC04, CSLC04, Cellulose-synthase-like C4, CSLC4, CELLULOSE-SYNTHASE LIKE C4 |
-0.281 |
-0.202 |
| 339 |
263192_at |
AT1G36160
|
ACC1, acetyl-CoA carboxylase 1, AT-ACC1, EMB22, EMBRYO DEFECTIVE 22, GK, GURKE, GSD1, GLOSSYHEAD 1, PAS3, PASTICCINO 3, SFR3, SENSITIVE TO FREEZING 3 |
-0.281 |
0.003 |
| 340 |
244997_at |
ATCG00170
|
RPOC2 |
-0.281 |
0.096 |
| 341 |
263126_at |
AT1G78460
|
[SOUL heme-binding family protein] |
-0.280 |
0.122 |
| 342 |
258386_at |
AT3G15520
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.280 |
0.124 |
| 343 |
262172_at |
AT1G74970
|
RPS9, ribosomal protein S9, TWN3 |
-0.279 |
-0.134 |
| 344 |
267380_at |
AT2G26170
|
CYP711A1, cytochrome P450, family 711, subfamily A, polypeptide 1, MAX1, MORE AXILLARY BRANCHES 1 |
-0.278 |
0.210 |
| 345 |
255277_at |
AT4G04890
|
PDF2, protodermal factor 2 |
-0.277 |
0.081 |
| 346 |
253921_at |
AT4G26900
|
AT-HF, HIS HF, HISN4 |
-0.275 |
-0.346 |
| 347 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.273 |
0.034 |
| 348 |
251969_at |
AT3G53130
|
CYP97C1, CYTOCHROME P450 97C1, LUT1, LUTEIN DEFICIENT 1 |
-0.272 |
-0.431 |
| 349 |
250812_at |
AT5G04900
|
NOL, NYC1-like |
-0.268 |
0.053 |