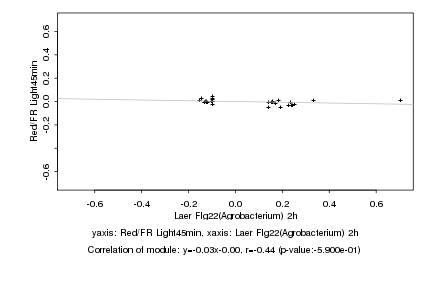
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Laer Flg22(Agrobacterium) 2h |
Log2 signal ratio
Red/FR Light45min |
| 1 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
0.702 |
0.009 |
| 2 |
250987_at |
AT5G02860
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.330 |
0.009 |
| 3 |
247786_at |
AT5G58600
|
PMR5, POWDERY MILDEW RESISTANT 5, TBL44, TRICHOME BIREFRINGENCE-LIKE 44 |
0.250 |
-0.020 |
| 4 |
250860_at |
AT5G04770
|
ATCAT6, ARABIDOPSIS THALIANA CATIONIC AMINO ACID TRANSPORTER 6, CAT6, cationic amino acid transporter 6 |
0.239 |
-0.026 |
| 5 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
0.238 |
-0.026 |
| 6 |
261937_at |
AT1G22570
|
[Major facilitator superfamily protein] |
0.231 |
-0.003 |
| 7 |
250194_at |
AT5G14550
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.222 |
-0.029 |
| 8 |
255931_at |
AT1G12710
|
AtPP2-A12, phloem protein 2-A12, PP2-A12, phloem protein 2-A12 |
0.191 |
-0.044 |
| 9 |
267048_at |
AT2G34200
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.181 |
0.017 |
| 10 |
254931_at |
AT4G11460
|
CRK30, cysteine-rich RLK (RECEPTOR-like protein kinase) 30 |
0.170 |
-0.010 |
| 11 |
253253_at |
AT4G34750
|
SAUR49, SMALL AUXIN UPREGULATED RNA 49 |
0.154 |
-0.000 |
| 12 |
251631_at |
AT3G57430
|
OTP84, ORGANELLE TRANSCRIPT PROCESSING 84 |
0.154 |
0.001 |
| 13 |
247641_at |
AT5G60540
|
ATPDX2, PYRIDOXINE BIOSYNTHESIS 2, EMB2407, EMBRYO DEFECTIVE 2407, PDX2, pyridoxine biosynthesis 2 |
0.152 |
-0.000 |
| 14 |
245190_at |
AT1G67690
|
[Zincin-like metalloproteases family protein] |
0.139 |
-0.044 |
| 15 |
257655_at |
AT3G13350
|
[HMG (high mobility group) box protein with ARID/BRIGHT DNA-binding domain] |
0.138 |
-0.004 |
| 16 |
263032_at |
AT1G23850
|
unknown |
-0.157 |
0.016 |
| 17 |
247354_at |
AT5G63590
|
ATFLS3, FLS3, flavonol synthase 3 |
-0.146 |
0.026 |
| 18 |
250855_at |
AT5G04730
|
[Ankyrin-repeat containing protein] |
-0.134 |
0.000 |
| 19 |
262998_at |
AT1G54280
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
-0.132 |
-0.001 |
| 20 |
261727_at |
AT1G76090
|
SMT3, sterol methyltransferase 3 |
-0.131 |
0.003 |
| 21 |
260948_at |
AT1G06100
|
[Fatty acid desaturase family protein] |
-0.127 |
0.002 |
| 22 |
251722_at |
AT3G56200
|
[Transmembrane amino acid transporter family protein] |
-0.124 |
-0.006 |
| 23 |
251688_at |
AT3G56480
|
[myosin heavy chain-related] |
-0.123 |
-0.003 |
| 24 |
256911_at |
AT3G24090
|
GFAT, L-glutamine D-fructose-6-phosphate amidotransferase |
-0.106 |
0.005 |
| 25 |
248352_at |
AT5G52300
|
LTI65, LOW-TEMPERATURE-INDUCED 65, RD29B, RESPONSIVE TO DESSICATION 29B |
-0.101 |
0.046 |
| 26 |
258817_at |
AT3G04750
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.101 |
0.001 |
| 27 |
253514_at |
AT4G31805
|
POLAR, POLAR LOCALIZATION DURING ASYMMETRIC DIVISION AND REDISTRIBUTION |
-0.101 |
0.018 |
| 28 |
254458_at |
AT4G21180
|
ATERDJ2B |
-0.100 |
0.028 |
| 29 |
254634_at |
AT4G18650
|
[transcription factor-related] |
-0.099 |
0.002 |
| 30 |
255666_at |
AT4G00390
|
[DNA-binding storekeeper protein-related transcriptional regulator] |
-0.098 |
-0.023 |