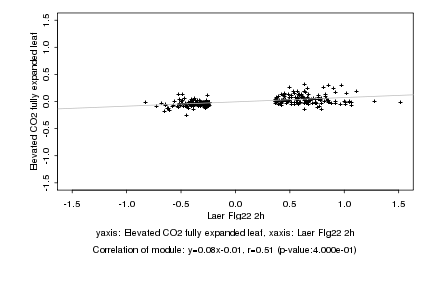
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Laer Flg22 2h |
Log2 signal ratio
Elevated CO2 fully expanded leaf |
| 1 |
253505_at |
AT4G31970
|
CYP82C2, cytochrome P450, family 82, subfamily C, polypeptide 2 |
1.513 |
-0.003 |
| 2 |
261475_at |
AT1G14550
|
[Peroxidase superfamily protein] |
1.275 |
0.006 |
| 3 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
1.112 |
0.189 |
| 4 |
261006_at |
AT1G26410
|
[FAD-binding Berberine family protein] |
1.063 |
-0.016 |
| 5 |
266784_at |
AT2G28960
|
[Leucine-rich repeat protein kinase family protein] |
1.060 |
-0.059 |
| 6 |
249586_at |
AT5G37840
|
unknown |
1.047 |
-0.010 |
| 7 |
255111_at |
AT4G08780
|
[Peroxidase superfamily protein] |
1.044 |
0.010 |
| 8 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
1.016 |
0.163 |
| 9 |
248090_at |
AT5G55090
|
MAPKKK15, mitogen-activated protein kinase kinase kinase 15 |
1.007 |
-0.042 |
| 10 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
1.004 |
-0.014 |
| 11 |
246349_at |
AT1G51915
|
[cryptdin protein-related] |
0.994 |
0.007 |
| 12 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.973 |
0.296 |
| 13 |
251743_at |
AT3G55890
|
[Yippee family putative zinc-binding protein] |
0.959 |
-0.037 |
| 14 |
251970_at |
AT3G53150
|
UGT73D1, UDP-glucosyl transferase 73D1 |
0.917 |
0.170 |
| 15 |
265796_at |
AT2G35730
|
[Heavy metal transport/detoxification superfamily protein ] |
0.915 |
-0.026 |
| 16 |
267588_at |
AT2G42060
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.914 |
0.012 |
| 17 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
0.896 |
0.256 |
| 18 |
250153_at |
AT5G15130
|
ATWRKY72, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 72, WRKY72, WRKY DNA-binding protein 72 |
0.881 |
-0.028 |
| 19 |
266976_at |
AT2G39410
|
[alpha/beta-Hydrolases superfamily protein] |
0.861 |
-0.007 |
| 20 |
267567_at |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
0.851 |
0.309 |
| 21 |
247205_at |
AT5G64890
|
PROPEP2, elicitor peptide 2 precursor |
0.850 |
0.033 |
| 22 |
247912_at |
AT5G57480
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.846 |
-0.003 |
| 23 |
250695_at |
AT5G06740
|
LecRK-S.5, L-type lectin receptor kinase S.5 |
0.844 |
0.020 |
| 24 |
265705_at |
AT2G03410
|
[Mo25 family protein] |
0.834 |
0.072 |
| 25 |
250267_at |
AT5G12930
|
unknown |
0.827 |
0.103 |
| 26 |
256933_at |
AT3G22600
|
LTPG5, glycosylphosphatidylinositol-anchored lipid protein transfer 5 |
0.820 |
0.144 |
| 27 |
260557_at |
AT2G43610
|
[Chitinase family protein] |
0.814 |
0.012 |
| 28 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
0.807 |
0.268 |
| 29 |
252153_at |
AT3G51360
|
[Eukaryotic aspartyl protease family protein] |
0.785 |
0.059 |
| 30 |
262831_at |
AT1G14730
|
[Cytochrome b561/ferric reductase transmembrane protein family] |
0.784 |
-0.129 |
| 31 |
247610_at |
AT5G60630
|
unknown |
0.784 |
-0.029 |
| 32 |
250415_at |
AT5G11210
|
ATGLR2.5, ARABIDOPSIS THALIANA GLU, GLR2.5, glutamate receptor 2.5 |
0.781 |
0.035 |
| 33 |
248528_at |
AT5G50760
|
SAUR55, SMALL AUXIN UPREGULATED RNA 55 |
0.779 |
0.045 |
| 34 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
0.766 |
-0.084 |
| 35 |
264766_at |
AT1G61420
|
[S-locus lectin protein kinase family protein] |
0.762 |
0.080 |
| 36 |
249057_at |
AT5G44480
|
DUR, DEFECTIVE UGE IN ROOT |
0.757 |
0.116 |
| 37 |
264280_at |
AT1G61820
|
BGLU46, beta glucosidase 46 |
0.754 |
0.053 |
| 38 |
266884_at |
AT2G44790
|
UCC2, uclacyanin 2 |
0.752 |
-0.092 |
| 39 |
252222_at |
AT3G49845
|
WIH3, WINDHOSE 3 |
0.741 |
-0.033 |
| 40 |
261429_at |
AT1G18860
|
ATWRKY61, WRKY DNA-BINDING PROTEIN 61, WRKY61, WRKY DNA-binding protein 61 |
0.735 |
-0.014 |
| 41 |
261842_at |
AT1G16160
|
WAKL5, wall associated kinase-like 5 |
0.733 |
0.050 |
| 42 |
252568_at |
AT3G45410
|
LecRK-I.3, L-type lectin receptor kinase I.3 |
0.732 |
-0.003 |
| 43 |
254347_at |
AT4G22070
|
ATWRKY31, WRKY31, WRKY DNA-binding protein 31 |
0.728 |
-0.031 |
| 44 |
253012_at |
AT4G37900
|
unknown |
0.709 |
0.064 |
| 45 |
262131_at |
AT1G02900
|
ATRALF1, RAPID ALKALINIZATION FACTOR 1, RALF1, rapid alkalinization factor 1, RALFL1, RALF-LIKE 1 |
0.701 |
-0.020 |
| 46 |
259439_at |
AT1G01480
|
ACS2, 1-amino-cyclopropane-1-carboxylate synthase 2, AT-ACC2 |
0.692 |
0.053 |
| 47 |
260432_at |
AT1G68150
|
ATWRKY9, WRKY9, WRKY DNA-binding protein 9 |
0.687 |
0.025 |
| 48 |
246228_at |
AT4G36430
|
[Peroxidase superfamily protein] |
0.683 |
0.021 |
| 49 |
247831_at |
AT5G58540
|
[Protein kinase superfamily protein] |
0.676 |
0.028 |
| 50 |
258075_at |
AT3G25900
|
ATHMT-1, HMT-1 |
0.667 |
-0.053 |
| 51 |
248700_at |
AT5G48400
|
ATGLR1.2, GLR1.2, GLUTAMATE RECEPTOR 1.2 |
0.667 |
-0.005 |
| 52 |
248176_at |
AT5G54650
|
ATFH5, FORMIN HOMOLOGY 5, Fh5, formin homology5 |
0.667 |
0.010 |
| 53 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
0.666 |
0.074 |
| 54 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
0.664 |
0.131 |
| 55 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
0.662 |
0.255 |
| 56 |
248701_at |
AT5G48410
|
ATGLR1.3, ARABIDOPSIS THALIANA GLUTAMATE RECEPTOR 1.3, GLR1.3, glutamate receptor 1.3 |
0.662 |
0.077 |
| 57 |
248464_at |
AT5G51160
|
[Ankyrin repeat family protein] |
0.650 |
-0.033 |
| 58 |
257583_at |
AT1G66480
|
[plastid movement impaired 2 (PMI2)] |
0.650 |
0.031 |
| 59 |
256378_at |
AT1G66830
|
[Leucine-rich repeat protein kinase family protein] |
0.639 |
-0.007 |
| 60 |
253178_at |
AT4G35170
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.636 |
0.019 |
| 61 |
250090_at |
AT5G17330
|
GAD, glutamate decarboxylase, GAD1, GLUTAMATE DECARBOXYLASE 1 |
0.635 |
0.181 |
| 62 |
265846_at |
AT2G35770
|
scpl28, serine carboxypeptidase-like 28 |
0.631 |
-0.021 |
| 63 |
261216_at |
AT1G33030
|
[O-methyltransferase family protein] |
0.631 |
0.315 |
| 64 |
260403_at |
AT1G69810
|
ATWRKY36, WRKY36, WRKY DNA-binding protein 36 |
0.631 |
0.064 |
| 65 |
256781_at |
AT3G13650
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.630 |
-0.142 |
| 66 |
251096_at |
AT5G01550
|
LecRK-VI.3, L-type lectin receptor kinase VI.3, LECRKA4.2, lectin receptor kinase a4.1 |
0.629 |
0.020 |
| 67 |
247618_at |
AT5G60280
|
LecRK-I.8, L-type lectin receptor kinase I.8 |
0.628 |
0.191 |
| 68 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.626 |
0.200 |
| 69 |
252448_at |
AT3G47050
|
[Glycosyl hydrolase family protein] |
0.622 |
0.081 |
| 70 |
258752_at |
AT3G09520
|
ATEXO70H4, exocyst subunit exo70 family protein H4, EXO70H4, exocyst subunit exo70 family protein H4 |
0.620 |
0.067 |
| 71 |
260040_at |
AT1G68765
|
IDA, INFLORESCENCE DEFICIENT IN ABSCISSION |
0.609 |
0.097 |
| 72 |
259443_at |
AT1G02360
|
[Chitinase family protein] |
0.606 |
0.075 |
| 73 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
0.605 |
0.147 |
| 74 |
267045_at |
AT2G34180
|
ATWL2, WPL4-LIKE 2, CIPK13, CBL-interacting protein kinase 13, SnRK3.7, SNF1-RELATED PROTEIN KINASE 3.7, WL2, WPL4-LIKE 2 |
0.590 |
-0.006 |
| 75 |
249364_at |
AT5G40590
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.590 |
-0.015 |
| 76 |
253173_at |
AT4G35110
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.586 |
0.135 |
| 77 |
267226_at |
AT2G44010
|
unknown |
0.585 |
-0.006 |
| 78 |
259297_at |
AT3G05360
|
AtRLP30, receptor like protein 30, RLP30, receptor like protein 30 |
0.583 |
0.118 |
| 79 |
258342_at |
AT3G22800
|
[Leucine-rich repeat (LRR) family protein] |
0.581 |
-0.026 |
| 80 |
266785_at |
AT2G28970
|
[Leucine-rich repeat protein kinase family protein] |
0.576 |
-0.038 |
| 81 |
266581_at |
AT2G46140
|
[Late embryogenesis abundant protein] |
0.571 |
0.131 |
| 82 |
253193_at |
AT4G35380
|
[SEC7-like guanine nucleotide exchange family protein] |
0.571 |
0.063 |
| 83 |
262303_at |
AT1G70920
|
ATHB18, homeobox-leucine zipper protein 18, HB18, homeobox-leucine zipper protein 18 |
0.568 |
-0.000 |
| 84 |
254468_at |
AT4G20460
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.568 |
0.031 |
| 85 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
0.567 |
0.193 |
| 86 |
262942_at |
AT1G79450
|
ALIS5, ALA-interacting subunit 5 |
0.565 |
0.076 |
| 87 |
254060_at |
AT4G25350
|
SHB1, SHORT HYPOCOTYL UNDER BLUE1 |
0.560 |
0.048 |
| 88 |
261815_at |
AT1G08320
|
bZIP21, TGA9, TGACG (TGA) motif-binding protein 9 |
0.559 |
-0.015 |
| 89 |
252939_at |
AT4G39230
|
[NmrA-like negative transcriptional regulator family protein] |
0.544 |
0.082 |
| 90 |
245089_at |
AT2G45290
|
TKL2, transketolase 2 |
0.542 |
-0.040 |
| 91 |
247297_at |
AT5G64100
|
[Peroxidase superfamily protein] |
0.535 |
-0.002 |
| 92 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
0.535 |
0.156 |
| 93 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
0.525 |
0.203 |
| 94 |
246912_at |
AT5G25820
|
[Exostosin family protein] |
0.515 |
0.081 |
| 95 |
248381_at |
AT5G51830
|
[pfkB-like carbohydrate kinase family protein] |
0.514 |
0.121 |
| 96 |
265994_at |
AT2G24170
|
[Endomembrane protein 70 protein family] |
0.509 |
-0.010 |
| 97 |
265355_at |
AT2G16760
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.508 |
-0.048 |
| 98 |
250640_at |
AT5G07150
|
[Leucine-rich repeat protein kinase family protein] |
0.492 |
0.033 |
| 99 |
252940_at |
AT4G39270
|
[Leucine-rich repeat protein kinase family protein] |
0.491 |
0.080 |
| 100 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
0.489 |
0.275 |
| 101 |
249860_at |
AT5G22860
|
[Serine carboxypeptidase S28 family protein] |
0.486 |
0.011 |
| 102 |
262444_at |
AT1G47480
|
[alpha/beta-Hydrolases superfamily protein] |
0.484 |
0.141 |
| 103 |
265306_at |
AT2G20320
|
[DENN (AEX-3) domain-containing protein] |
0.482 |
0.020 |
| 104 |
248129_at |
AT5G54780
|
[Ypt/Rab-GAP domain of gyp1p superfamily protein] |
0.470 |
0.004 |
| 105 |
256321_at |
AT1G55020
|
ATLOX1, ARABIDOPSIS LIPOXYGENASE 1, LOX1, lipoxygenase 1 |
0.468 |
-0.036 |
| 106 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
0.458 |
0.042 |
| 107 |
245300_at |
AT4G16350
|
CBL6, calcineurin B-like protein 6, SCABP2, SOS3-LIKE CALCIUM BINDING PROTEIN 2 |
0.457 |
0.036 |
| 108 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
0.448 |
0.034 |
| 109 |
254571_at |
AT4G19370
|
unknown |
0.446 |
0.139 |
| 110 |
265214_at |
AT1G05000
|
AtPFA-DSP1, PFA-DSP1, plant and fungi atypical dual-specificity phosphatase 1 |
0.445 |
0.093 |
| 111 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.443 |
0.163 |
| 112 |
248286_at |
AT5G52870
|
MAKR5, MEMBRANE-ASSOCIATED KINASE REGULATOR 5 |
0.439 |
0.042 |
| 113 |
259975_at |
AT1G76470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.438 |
0.113 |
| 114 |
267207_at |
AT2G30840
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.432 |
0.031 |
| 115 |
266669_at |
AT2G29750
|
UGT71C1, UDP-glucosyl transferase 71C1 |
0.427 |
0.009 |
| 116 |
252964_at |
AT4G38830
|
CRK26, cysteine-rich RLK (RECEPTOR-like protein kinase) 26 |
0.425 |
0.011 |
| 117 |
259149_at |
AT3G10340
|
PAL4, phenylalanine ammonia-lyase 4 |
0.422 |
-0.069 |
| 118 |
247229_at |
AT5G65160
|
TPR14, tetratricopeptide repeat 14 |
0.420 |
-0.008 |
| 119 |
267548_at |
AT2G32660
|
AtRLP22, receptor like protein 22, RLP22, receptor like protein 22 |
0.417 |
-0.029 |
| 120 |
256664_at |
AT3G12040
|
[DNA-3-methyladenine glycosylase (MAG)] |
0.416 |
0.023 |
| 121 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
0.415 |
0.138 |
| 122 |
245929_at |
AT5G24760
|
[GroES-like zinc-binding dehydrogenase family protein] |
0.410 |
-0.031 |
| 123 |
249372_at |
AT5G40760
|
G6PD6, glucose-6-phosphate dehydrogenase 6 |
0.399 |
0.035 |
| 124 |
258557_at |
AT3G05990
|
[Leucine-rich repeat (LRR) family protein] |
0.395 |
-0.051 |
| 125 |
258002_at |
AT3G28930
|
AIG2, AVRRPT2-INDUCED GENE 2 |
0.387 |
0.102 |
| 126 |
267518_at |
AT2G30500
|
NET4B, Networked 4B |
0.386 |
0.027 |
| 127 |
259309_at |
AT3G05050
|
[Protein kinase superfamily protein] |
0.381 |
0.016 |
| 128 |
249456_at |
AT5G39410
|
[Saccharopine dehydrogenase ] |
0.381 |
0.002 |
| 129 |
250997_at |
AT5G02570
|
[Histone superfamily protein] |
0.380 |
0.014 |
| 130 |
249783_at |
AT5G24270
|
ATSOS3, SALT OVERLY SENSITIVE 3, CBL4, CALCINEURIN B-LIKE PROTEIN 4, SOS3, SALT OVERLY SENSITIVE 3 |
0.376 |
0.078 |
| 131 |
264574_at |
AT1G05300
|
ZIP5, zinc transporter 5 precursor |
0.376 |
0.005 |
| 132 |
258287_at |
AT3G15990
|
SULTR3;4, sulfate transporter 3;4 |
0.375 |
-0.017 |
| 133 |
267544_at |
AT2G32720
|
ATCB5-B, ARABIDOPSIS CYTOCHROME B5 ISOFORM B, B5 #4, CB5-B, cytochrome B5 isoform B, CYTB5-D |
0.372 |
0.001 |
| 134 |
253292_at |
AT4G33985
|
unknown |
0.372 |
0.049 |
| 135 |
252121_at |
AT3G51160
|
GMD2, GDP-D-MANNOSE-4,6-DEHYDRATASE 2, MUR1, MURUS 1, MUR_1, MURUS 1 |
0.370 |
0.062 |
| 136 |
248410_at |
AT5G51570
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
0.369 |
0.034 |
| 137 |
258691_at |
AT3G08630
|
unknown |
0.367 |
-0.019 |
| 138 |
260392_at |
AT1G74030
|
ENO1, enolase 1 |
0.364 |
0.008 |
| 139 |
262457_at |
AT1G11200
|
unknown |
0.363 |
0.009 |
| 140 |
252437_at |
AT3G47380
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.363 |
0.052 |
| 141 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
-0.834 |
-0.015 |
| 142 |
245980_at |
AT5G13140
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.726 |
-0.074 |
| 143 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
-0.687 |
-0.025 |
| 144 |
254794_at |
AT4G12970
|
EPFL9, STOMAGEN, STOMAGEN |
-0.653 |
-0.175 |
| 145 |
247151_at |
AT5G65640
|
bHLH093, beta HLH protein 93 |
-0.648 |
-0.046 |
| 146 |
246275_at |
AT4G36540
|
BEE2, BR enhanced expression 2 |
-0.645 |
-0.062 |
| 147 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.627 |
-0.128 |
| 148 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
-0.622 |
-0.113 |
| 149 |
247882_at |
AT5G57785
|
unknown |
-0.615 |
-0.148 |
| 150 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.582 |
-0.090 |
| 151 |
248075_at |
AT5G55740
|
CRR21, chlororespiratory reduction 21 |
-0.575 |
-0.068 |
| 152 |
263482_at |
AT2G03980
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.567 |
0.006 |
| 153 |
253650_at |
AT4G30020
|
[PA-domain containing subtilase family protein] |
-0.534 |
-0.078 |
| 154 |
266778_at |
AT2G29090
|
CYP707A2, cytochrome P450, family 707, subfamily A, polypeptide 2 |
-0.532 |
0.144 |
| 155 |
265377_at |
AT2G05790
|
[O-Glycosyl hydrolases family 17 protein] |
-0.528 |
-0.097 |
| 156 |
263386_at |
AT2G40150
|
TBL28, TRICHOME BIREFRINGENCE-LIKE 28 |
-0.520 |
-0.035 |
| 157 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
-0.517 |
0.047 |
| 158 |
266790_at |
AT2G28950
|
ATEXP6, ARABIDOPSIS THALIANA TEXPANSIN 6, ATEXPA6, expansin A6, ATHEXP ALPHA 1.8, EXPA6, expansin A6 |
-0.517 |
-0.042 |
| 159 |
252168_at |
AT3G50440
|
ATMES10, ARABIDOPSIS THALIANA METHYL ESTERASE 10, MES10, methyl esterase 10 |
-0.510 |
-0.062 |
| 160 |
254280_at |
AT4G22756
|
ATSMO1-2, SMO1-2, sterol C4-methyl oxidase 1-2 |
-0.499 |
0.007 |
| 161 |
264611_at |
AT1G04680
|
[Pectin lyase-like superfamily protein] |
-0.489 |
-0.036 |
| 162 |
261927_at |
AT1G22500
|
AtATL15, Arabidopsis thaliana Arabidopsis toxicos en levadura 15, ATL15, Arabidopsis toxicos en levadura 15 |
-0.488 |
0.144 |
| 163 |
256578_at |
AT3G28200
|
[Peroxidase superfamily protein] |
-0.487 |
0.021 |
| 164 |
262569_at |
AT1G15180
|
[MATE efflux family protein] |
-0.484 |
-0.088 |
| 165 |
257129_at |
AT3G20100
|
CYP705A19, cytochrome P450, family 705, subfamily A, polypeptide 19 |
-0.474 |
0.056 |
| 166 |
262753_at |
AT1G16340
|
ATKDSA2, ATKSDA |
-0.466 |
-0.036 |
| 167 |
259207_at |
AT3G09050
|
unknown |
-0.464 |
-0.048 |
| 168 |
256792_at |
AT3G22150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.462 |
-0.080 |
| 169 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.457 |
-0.257 |
| 170 |
259658_at |
AT1G55370
|
NDF5, NDH-dependent cyclic electron flow 5 |
-0.454 |
-0.064 |
| 171 |
264672_at |
AT1G09750
|
[Eukaryotic aspartyl protease family protein] |
-0.450 |
-0.077 |
| 172 |
246487_at |
AT5G16030
|
unknown |
-0.444 |
-0.090 |
| 173 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
-0.438 |
-0.004 |
| 174 |
259308_at |
AT3G05180
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.433 |
-0.119 |
| 175 |
257717_at |
AT3G18390
|
EMB1865, embryo defective 1865 |
-0.426 |
-0.042 |
| 176 |
250975_at |
AT5G03050
|
unknown |
-0.421 |
0.004 |
| 177 |
253317_at |
AT4G33960
|
unknown |
-0.410 |
0.052 |
| 178 |
247377_at |
AT5G63180
|
[Pectin lyase-like superfamily protein] |
-0.409 |
-0.055 |
| 179 |
258528_at |
AT3G06770
|
[Pectin lyase-like superfamily protein] |
-0.406 |
-0.089 |
| 180 |
251317_at |
AT3G61490
|
[Pectin lyase-like superfamily protein] |
-0.406 |
-0.048 |
| 181 |
264315_at |
AT1G70370
|
PG2, polygalacturonase 2 |
-0.402 |
-0.030 |
| 182 |
248095_at |
AT5G55230
|
ATMAP65-1, microtubule-associated proteins 65-1, MAP65-1, MAP65-1, microtubule-associated proteins 65-1 |
-0.402 |
-0.015 |
| 183 |
259763_at |
AT1G77630
|
LYM3, lysin-motif (LysM) domain protein 3, LYP3, LysM-containing receptor protein 3 |
-0.402 |
-0.041 |
| 184 |
264247_at |
AT1G60160
|
[Potassium transporter family protein] |
-0.401 |
-0.053 |
| 185 |
267472_at |
AT2G02850
|
ARPN, plantacyanin |
-0.400 |
-0.051 |
| 186 |
247746_at |
AT5G58970
|
ATUCP2, uncoupling protein 2, UCP2, uncoupling protein 2 |
-0.400 |
-0.031 |
| 187 |
259021_at |
AT3G07540
|
[Actin-binding FH2 (formin homology 2) family protein] |
-0.389 |
-0.079 |
| 188 |
254333_at |
AT4G22753
|
ATSMO1-3, SMO1-3, sterol 4-alpha methyl oxidase 1-3 |
-0.388 |
0.022 |
| 189 |
262730_at |
AT1G16390
|
ATOCT3, organic cation/carnitine transporter 3, OCT3, organic cation/carnitine transporter 3 |
-0.387 |
-0.042 |
| 190 |
252215_at |
AT3G50240
|
KICP-02 |
-0.386 |
-0.133 |
| 191 |
261522_at |
AT1G71710
|
[DNAse I-like superfamily protein] |
-0.384 |
-0.058 |
| 192 |
266996_at |
AT2G34490
|
CYP710A2, cytochrome P450, family 710, subfamily A, polypeptide 2 |
-0.384 |
0.067 |
| 193 |
263207_at |
AT1G10550
|
XET, XYLOGLUCAN:XYLOGLUCOSYL TRANSFERASE 33, XTH33, xyloglucan:xyloglucosyl transferase 33 |
-0.376 |
0.035 |
| 194 |
259958_at |
AT1G53730
|
SRF6, STRUBBELIG-receptor family 6 |
-0.368 |
-0.042 |
| 195 |
250884_at |
AT5G03940
|
54CP, 54 CHLOROPLAST PROTEIN, CPSRP54, chloroplast signal recognition particle 54 kDa subunit, FFC, FIFTY-FOUR CHLOROPLAST HOMOLOGUE, SRP54CP, SIGNAL RECOGNITION PARTICLE 54 KDA SUBUNIT CHLOROPLAST PROTEIN |
-0.364 |
-0.044 |
| 196 |
253286_at |
AT4G34260
|
AXY8, ALTERED XYLOGLUCAN 8, FUC95A |
-0.363 |
-0.036 |
| 197 |
255617_at |
AT4G01330
|
[Protein kinase superfamily protein] |
-0.363 |
-0.049 |
| 198 |
266635_at |
AT2G35470
|
unknown |
-0.360 |
-0.016 |
| 199 |
259812_at |
AT1G49840
|
unknown |
-0.360 |
-0.014 |
| 200 |
253635_at |
AT4G30620
|
[Uncharacterised BCR, YbaB family COG0718] |
-0.358 |
-0.038 |
| 201 |
266802_at |
AT2G22900
|
[Galactosyl transferase GMA12/MNN10 family protein] |
-0.355 |
-0.048 |
| 202 |
249784_at |
AT5G24280
|
GMI1, GAMMA-IRRADIATION AND MITOMYCIN C INDUCED 1 |
-0.355 |
-0.010 |
| 203 |
258389_at |
AT3G15390
|
SDE5, silencing defective 5 |
-0.354 |
0.019 |
| 204 |
258055_at |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
-0.353 |
-0.047 |
| 205 |
260969_at |
AT1G12240
|
ATBETAFRUCT4, AtFRUCT4, AtVI2, FRUCT4, fructosidase 4, VAC-INV, VACUOLAR INVERTASE, VI2, vacuolar invertase 2 |
-0.348 |
-0.056 |
| 206 |
251899_at |
AT3G54400
|
[Eukaryotic aspartyl protease family protein] |
-0.346 |
-0.075 |
| 207 |
254911_at |
AT4G11100
|
unknown |
-0.344 |
-0.047 |
| 208 |
260774_at |
AT1G78290
|
SNRK2-8, SNF1-RELATED PROTEIN KINASE 2-8, SNRK2.8, SNF1-RELATED PROTEIN KINASE 2.8, SRK2C, SNF1-RELATED PROTEIN KINASE 2C |
-0.335 |
-0.007 |
| 209 |
255035_at |
AT4G09550
|
ATGIP1, ARABIDOPSIS ATGCP3 INTERACTING PROTEIN 1, GIP1, AtGCP3 interacting protein 1, GIP1a, GCP3-Interacting Protein 1a |
-0.334 |
0.019 |
| 210 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
-0.334 |
-0.038 |
| 211 |
261117_at |
AT1G75310
|
AUL1, auxilin-like 1 |
-0.334 |
-0.014 |
| 212 |
251169_at |
AT3G63210
|
MARD1, MEDIATOR OF ABA-REGULATED DORMANCY 1 |
-0.333 |
-0.053 |
| 213 |
258368_at |
AT3G14240
|
[Subtilase family protein] |
-0.331 |
-0.033 |
| 214 |
245426_at |
AT4G17540
|
unknown |
-0.330 |
-0.060 |
| 215 |
262648_at |
AT1G14030
|
LSMT-L, lysine methyltransferase (LSMT)-like |
-0.328 |
-0.050 |
| 216 |
267078_at |
AT2G40960
|
[Single-stranded nucleic acid binding R3H protein] |
-0.323 |
0.001 |
| 217 |
267158_at |
AT2G37640
|
ATEXP3, EXPANSIN 3, ATEXPA3, ARABIDOPSIS THALIANA EXPANSIN A3, ATHEXP ALPHA 1.9, EXP3 |
-0.322 |
-0.088 |
| 218 |
250669_at |
AT5G06870
|
ATPGIP2, ARABIDOPSIS POLYGALACTURONASE INHIBITING PROTEIN 2, PGIP2, polygalacturonase inhibiting protein 2 |
-0.321 |
-0.006 |
| 219 |
263322_at |
AT2G04270
|
RNE, RNase E, RNEE/G, RNAse E/G-like |
-0.317 |
-0.067 |
| 220 |
265570_at |
AT2G28310
|
unknown |
-0.314 |
-0.009 |
| 221 |
251084_at |
AT5G01520
|
AIRP2, ABA Insensitive RING Protein 2, AtAIRP2 |
-0.309 |
-0.011 |
| 222 |
250421_at |
AT5G11270
|
OCP3, overexpressor of cationic peroxidase 3 |
-0.309 |
-0.042 |
| 223 |
264507_at |
AT1G09415
|
NIMIN-3, NIM1-interacting 3 |
-0.302 |
-0.029 |
| 224 |
264224_at |
AT1G67440
|
emb1688, embryo defective 1688 |
-0.299 |
-0.057 |
| 225 |
263496_at |
AT2G42570
|
TBL39, TRICHOME BIREFRINGENCE-LIKE 39 |
-0.296 |
-0.004 |
| 226 |
266957_at |
AT2G34640
|
HMR, HEMERA, PTAC12, plastid transcriptionally active 12, TAC12 |
-0.294 |
-0.005 |
| 227 |
250540_at |
AT5G08580
|
[Calcium-binding EF hand family protein] |
-0.292 |
0.000 |
| 228 |
251278_at |
AT3G61780
|
emb1703, embryo defective 1703 |
-0.291 |
-0.072 |
| 229 |
257171_at |
AT3G23760
|
unknown |
-0.291 |
-0.078 |
| 230 |
252548_at |
AT3G45850
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.289 |
-0.093 |
| 231 |
253440_at |
AT4G32570
|
TIFY8, TIFY domain protein 8 |
-0.288 |
-0.067 |
| 232 |
248402_at |
AT5G52100
|
CRR1, chlororespiration reduction 1 |
-0.287 |
-0.055 |
| 233 |
260465_at |
AT1G10910
|
EMB3103, EMBRYO DEFECTIVE 3103 |
-0.286 |
-0.053 |
| 234 |
252184_at |
AT3G50660
|
AtDWF4, CLM, CLOMAZONE-RESISTANT, CYP90B1, CYTOCHROME P450 90B1, DWF4, DWARF 4, PSC1, PARTIALLY SUPPRESSING COI1 INSENSITIVITY TO JA 1, SAV1, SHADE AVOIDANCE 1, SNP2, SUPPRESSOR OF NPH4 2 |
-0.284 |
-0.031 |
| 235 |
252922_at |
AT4G39040
|
[RNA-binding CRS1 / YhbY (CRM) domain protein] |
-0.284 |
-0.033 |
| 236 |
262728_at |
AT1G75820
|
ATCLV1, CLV1, CLAVATA 1, FAS3, FASCIATA 3, FLO5, FLOWER DEVELOPMENT 5 |
-0.283 |
-0.011 |
| 237 |
261801_at |
AT1G30520
|
AAE14, acyl-activating enzyme 14 |
-0.282 |
-0.124 |
| 238 |
260109_at |
AT1G63260
|
TET10, tetraspanin10 |
-0.282 |
-0.022 |
| 239 |
253686_at |
AT4G29750
|
[CRS1 / YhbY (CRM) domain-containing protein] |
-0.276 |
-0.011 |
| 240 |
257573_at |
AT2G33990
|
iqd9, IQ-domain 9 |
-0.276 |
-0.043 |
| 241 |
256222_at |
AT1G56210
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.275 |
-0.078 |
| 242 |
261016_at |
AT1G26560
|
BGLU40, beta glucosidase 40 |
-0.274 |
-0.089 |
| 243 |
267586_at |
AT2G41950
|
unknown |
-0.274 |
-0.028 |
| 244 |
264476_at |
AT1G77130
|
GUX3, glucuronic acid substitution of xylan 3, PGSIP2, plant glycogenin-like starch initiation protein 2 |
-0.274 |
-0.011 |
| 245 |
248861_at |
AT5G46700
|
TET1, TETRASPANIN 1, TRN2, TORNADO 2 |
-0.273 |
-0.074 |
| 246 |
252852_at |
AT4G39900
|
unknown |
-0.273 |
-0.085 |
| 247 |
252189_at |
AT3G50070
|
CYCD3;3, CYCLIN D3;3 |
-0.272 |
0.010 |
| 248 |
255487_at |
AT4G02610
|
[Aldolase-type TIM barrel family protein] |
-0.272 |
-0.000 |
| 249 |
251227_at |
AT3G62700
|
ABCC14, ATP-binding cassette C14, ATMRP10, multidrug resistance-associated protein 10, MRP10, multidrug resistance-associated protein 10 |
-0.270 |
-0.106 |
| 250 |
258923_at |
AT3G10450
|
SCPL7, serine carboxypeptidase-like 7 |
-0.268 |
0.034 |
| 251 |
254328_at |
AT4G22570
|
APT3, adenine phosphoribosyl transferase 3 |
-0.268 |
-0.002 |
| 252 |
264963_at |
AT1G60600
|
ABC4, ABERRANT CHLOROPLAST DEVELOPMENT 4 |
-0.267 |
-0.054 |
| 253 |
245138_at |
AT2G45190
|
AFO, ABNORMAL FLORAL ORGANS, FIL, FILAMENTOUS FLOWER, YAB1, YABBY1 |
-0.265 |
-0.070 |
| 254 |
258475_at |
AT3G02660
|
EMB2768, EMBRYO DEFECTIVE 2768, FAC31, EMBRYONIC FACTOR 31 |
-0.264 |
-0.046 |
| 255 |
245368_at |
AT4G15510
|
PPD1, PsbP-Domain Protein1 |
-0.264 |
-0.054 |
| 256 |
246200_at |
AT4G37240
|
unknown |
-0.263 |
-0.053 |
| 257 |
257856_at |
AT3G12930
|
[Lojap-related protein] |
-0.262 |
-0.043 |
| 258 |
250483_at |
AT5G10300
|
AtHNL, ATMES5, ARABIDOPSIS THALIANA METHYL ESTERASE 5, HNL, HYDROXYNITRILE LYASE, MES5, methyl esterase 5 |
-0.262 |
0.118 |
| 259 |
247348_at |
AT5G63810
|
AtBGAL10, Arabidopsis thaliana beta-galactosidase 10, BGAL10, beta-galactosidase 10 |
-0.261 |
-0.057 |
| 260 |
267562_at |
AT2G39670
|
[Radical SAM superfamily protein] |
-0.260 |
-0.019 |
| 261 |
264442_at |
AT1G27480
|
[alpha/beta-Hydrolases superfamily protein] |
-0.260 |
-0.042 |
| 262 |
265305_at |
AT2G20340
|
AAS, aromatic aldehyde synthase, AtAAS |
-0.258 |
-0.019 |
| 263 |
263591_at |
AT2G01910
|
ATMAP65-6, MAP65-6 |
-0.257 |
-0.025 |
| 264 |
248913_at |
AT5G45760
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.257 |
-0.006 |
| 265 |
259300_at |
AT3G05100
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.257 |
-0.034 |
| 266 |
265682_at |
AT2G24390
|
[AIG2-like (avirulence induced gene) family protein] |
-0.256 |
-0.002 |
| 267 |
252934_at |
AT4G39120
|
HISN7, HISTIDINE BIOSYNTHESIS 7, IMPL2, myo-inositol monophosphatase like 2 |
-0.256 |
-0.045 |
| 268 |
260548_at |
AT2G43360
|
BIO2, BIOTIN AUXOTROPH 2, BIOB, BIOTIN AUXOTROPH B |
-0.256 |
-0.068 |
| 269 |
263902_at |
AT2G36230
|
APG10, ALBINO AND PALE GREEN 10, HISN3 |
-0.255 |
-0.047 |
| 270 |
258324_at |
AT3G22780
|
ATTSO1, TSO1, CHINESE FOR 'UGLY' |
-0.255 |
-0.051 |
| 271 |
261695_at |
AT1G08520
|
ALB-1V, ALB1, ALBINA 1, CHLD, PDE166, PIGMENT DEFECTIVE EMBRYO 166, V157 |
-0.254 |
-0.030 |
| 272 |
258136_at |
AT3G24560
|
RSY3, RASPBERRY 3 |
-0.253 |
-0.090 |
| 273 |
260877_at |
AT1G21500
|
unknown |
-0.249 |
-0.063 |
| 274 |
249826_at |
AT5G23310
|
FSD3, Fe superoxide dismutase 3 |
-0.248 |
-0.006 |
| 275 |
247098_at |
AT5G66470
|
[RNA binding] |
-0.247 |
-0.014 |
| 276 |
261479_at |
AT1G14380
|
IQD28, IQ-domain 28 |
-0.247 |
-0.016 |
| 277 |
261796_at |
AT1G30440
|
[Phototropic-responsive NPH3 family protein] |
-0.246 |
-0.017 |
| 278 |
267013_at |
AT2G39180
|
ATCRR2, CCR2, CRINKLY4 related 2 |
-0.245 |
-0.008 |
| 279 |
257908_at |
AT3G25410
|
[Sodium Bile acid symporter family] |
-0.245 |
-0.065 |
| 280 |
260009_at |
AT1G67950
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.245 |
-0.010 |