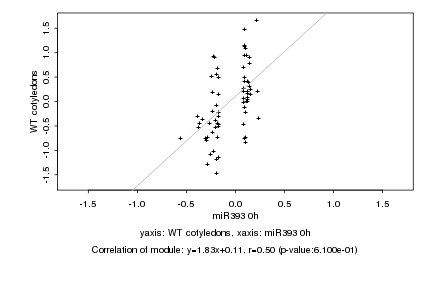
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
miR393 0h |
Log2 signal ratio
WT cotyledons |
| 1 |
250955_at |
AT5G03190
|
CPUORF47, conserved peptide upstream open reading frame 47 |
0.227 |
-0.344 |
| 2 |
251174_at |
AT3G63200
|
PLA IIIB, PLP9, PATATIN-like protein 9 |
0.216 |
0.220 |
| 3 |
252971_at |
AT4G38770
|
ATPRP4, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 4, PRP4, proline-rich protein 4 |
0.212 |
1.679 |
| 4 |
261819_at |
AT1G11410
|
[S-locus lectin protein kinase family protein] |
0.151 |
0.160 |
| 5 |
254737_at |
AT4G13840
|
CER26, ECERIFERUM 26 |
0.145 |
0.249 |
| 6 |
251853_at |
AT3G54790
|
[ARM repeat superfamily protein] |
0.137 |
0.317 |
| 7 |
260547_at |
AT2G43550
|
[Scorpion toxin-like knottin superfamily protein] |
0.135 |
0.906 |
| 8 |
247439_at |
AT5G62670
|
AHA11, H(+)-ATPase 11, HA11, H(+)-ATPase 11 |
0.134 |
0.797 |
| 9 |
262230_at |
AT1G68560
|
ATXYL1, ALPHA-XYLOSIDASE 1, AXY3, altered xyloglucan 3, TRG1, thermoinhibition resistant germination 1, XYL1, alpha-xylosidase 1 |
0.130 |
0.408 |
| 10 |
253218_at |
AT4G34980
|
SLP2, subtilisin-like serine protease 2 |
0.121 |
0.177 |
| 11 |
256890_at |
AT3G23830
|
AtGRP4, GR-RBP4, GRP4, glycine-rich RNA-binding protein 4 |
0.120 |
0.221 |
| 12 |
259005_at |
AT3G01930
|
[Major facilitator superfamily protein] |
0.117 |
0.056 |
| 13 |
259430_at |
AT1G01610
|
ATGPAT4, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 4, GPAT4, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 4 |
0.116 |
0.004 |
| 14 |
249986_at |
AT5G18460
|
unknown |
0.115 |
0.428 |
| 15 |
260857_at |
AT1G21880
|
LYM1, lysm domain GPI-anchored protein 1 precursor, LYP2, LysM-containing receptor protein 2 |
0.114 |
0.094 |
| 16 |
258543_at |
AT3G06870
|
[proline-rich family protein] |
0.111 |
0.004 |
| 17 |
249785_at |
AT5G24300
|
ATSS1, STARCH SYNTHASE 1, SS1, starch synthase 1 |
0.104 |
0.947 |
| 18 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
0.099 |
-0.213 |
| 19 |
251301_at |
AT3G61880
|
CYP78A9, cytochrome p450 78a9 |
0.098 |
-0.834 |
| 20 |
260288_at |
AT1G80530
|
[Major facilitator superfamily protein] |
0.096 |
1.097 |
| 21 |
258738_at |
AT3G05750
|
TRM6, TON1 Recruiting Motif 6 |
0.093 |
-0.718 |
| 22 |
249818_at |
AT5G23860
|
TUB8, tubulin beta 8 |
0.091 |
1.163 |
| 23 |
256681_at |
AT3G52340
|
ATSPP2, SUCROSE-PHOSPHATASE 2, SPP2, sucrose-6F-phosphate phosphohydrolase 2 |
0.090 |
1.143 |
| 24 |
258086_at |
AT3G25860
|
LTA2, PLE2, PLASTID E2 SUBUNIT OF PYRUVATE DECARBOXYLASE |
0.090 |
0.495 |
| 25 |
245417_at |
AT4G17360
|
[Formyl transferase] |
0.086 |
0.953 |
| 26 |
262582_at |
AT1G15410
|
[aspartate-glutamate racemase family] |
0.085 |
0.423 |
| 27 |
261903_at |
AT1G80740
|
CMT1, chromomethylase 1, DMT4, DNA METHYLTRANSFERASE 4 |
0.085 |
-0.750 |
| 28 |
263755_at |
AT2G21340
|
[MATE efflux family protein] |
0.084 |
1.493 |
| 29 |
264943_at |
AT1G76910
|
unknown |
0.083 |
-0.113 |
| 30 |
262870_at |
AT1G64710
|
[GroES-like zinc-binding dehydrogenase family protein] |
0.081 |
0.711 |
| 31 |
248028_at |
AT5G55620
|
unknown |
0.079 |
0.284 |
| 32 |
262920_at |
AT1G79500
|
AtkdsA1 |
0.078 |
-0.457 |
| 33 |
258185_at |
AT3G21580
|
ABCI12, ATP-binding cassette I12 |
0.076 |
0.081 |
| 34 |
250329_at |
AT5G11800
|
ATKEA6, ARABIDOPSIS THALIANA K+ EFFLUX ANTIPORTER 6, KEA6, K+ efflux antiporter 6 |
0.074 |
-0.019 |
| 35 |
266254_at |
AT2G27810
|
ATNAT12, ARABIDOPSIS NUCLEOBASE-ASCORBATE TRANSPORTER 12, NAT12, nucleobase-ascorbate transporter 12 |
0.074 |
0.206 |
| 36 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
-0.566 |
-0.751 |
| 37 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
-0.388 |
-0.292 |
| 38 |
253044_at |
AT4G37290
|
unknown |
-0.386 |
-0.512 |
| 39 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
-0.374 |
-0.434 |
| 40 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
-0.343 |
-0.363 |
| 41 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
-0.308 |
-0.751 |
| 42 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
-0.302 |
-0.798 |
| 43 |
246197_at |
AT4G37010
|
CEN2, centrin 2 |
-0.293 |
-1.285 |
| 44 |
260261_at |
AT1G68450
|
PDE337, PIGMENT DEFECTIVE 337 |
-0.289 |
-0.735 |
| 45 |
258203_at |
AT3G13950
|
unknown |
-0.265 |
-0.442 |
| 46 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
-0.256 |
-1.067 |
| 47 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
-0.246 |
0.520 |
| 48 |
245082_at |
AT2G23270
|
unknown |
-0.241 |
-0.198 |
| 49 |
258075_at |
AT3G25900
|
ATHMT-1, HMT-1 |
-0.240 |
0.200 |
| 50 |
256306_at |
AT1G30370
|
DLAH, DAD1-like acylhydrolase |
-0.237 |
-0.619 |
| 51 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
-0.233 |
-1.010 |
| 52 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
-0.231 |
0.929 |
| 53 |
264343_at |
AT1G11850
|
unknown |
-0.218 |
0.914 |
| 54 |
255630_at |
AT4G00700
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
-0.212 |
-0.522 |
| 55 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
-0.206 |
-0.373 |
| 56 |
256883_at |
AT3G26440
|
unknown |
-0.203 |
0.555 |
| 57 |
264433_at |
AT1G61810
|
BGLU45, beta-glucosidase 45 |
-0.202 |
-1.181 |
| 58 |
250493_at |
AT5G09800
|
[ARM repeat superfamily protein] |
-0.198 |
-1.466 |
| 59 |
248154_at |
AT5G54400
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.194 |
-0.073 |
| 60 |
253754_at |
AT4G29020
|
[glycine-rich protein] |
-0.192 |
0.695 |
| 61 |
260460_at |
AT1G68230
|
[Reticulon family protein] |
-0.189 |
-0.450 |
| 62 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
-0.187 |
-0.733 |
| 63 |
261175_at |
AT1G04800
|
[glycine-rich protein] |
-0.182 |
0.494 |
| 64 |
252825_at |
AT4G39890
|
AtRABH1c, RAB GTPase homolog H1C, RABH1c, RAB GTPase homolog H1C |
-0.182 |
-0.451 |
| 65 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
-0.182 |
-0.503 |
| 66 |
256933_at |
AT3G22600
|
LTPG5, glycosylphosphatidylinositol-anchored lipid protein transfer 5 |
-0.181 |
0.159 |
| 67 |
250796_at |
AT5G05300
|
unknown |
-0.181 |
-0.206 |
| 68 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
-0.178 |
-0.300 |
| 69 |
247333_at |
AT5G63600
|
ATFLS5, FLS5, flavonol synthase 5 |
-0.178 |
-1.126 |